Gene
KWMTBOMO04221
Pre Gene Modal
BGIBMGA014260
Annotation
PREDICTED:_metabotropic_glutamate_receptor_2-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.836
Sequence
CDS
ATGAGTGCGGCGGCTCGCGTGCTGCTGGCGACGCTGCTGGCGACGGCGGCCCTCGCCTCGCACGCTGACCGCACCCACCTCGCCAACATCAGCTCCGCTCTGACCAACCCCACGACTCAGCTGTTCTATGATGACTTGCTTCCTTCTCATATACTCAGAAGTAAAAGAAACGCACACACCCTCATTGCCACTTTCGATATTTCAAAGATCTTCAAGAGAAAGAAGCTGCAGAGCAAGCAAGGGGACTCGCCAAACCCTACTCCCTCCTCCCTCCTCTCCGACAACCTGTCCGCCAGTATGGTCGCTGAGCTGCTGGCCACGAGCCTCGCCAATGTGTCTCTGGTCGTCGACACCACTCCACTCACTGATCCGATAGAACTCACCACTTCACTGGTCTCAAACAGTACTATCTCTACGAAAAGAGTCTCTACACCCTTCACACAAATTTCGAATCCGAAAAATTCCTTGAAACTTAAAAGTAAAAGTGAATCGAAGGGTGTCGATGCTAGCGTGTCGTCACGAGTTGGAGTCAAGAAGAAGGTATGGGAGAACGAGACGGTGTGGCCCGTGAAGCACGCCGCCGTTGTCGACGGAGACATCATCCTCGGCGGACTCATGATGGTCCACCGTCGGTCGGAGGTGGCGACGTGCGGGCCAGTGATGGCGCAGGGCGGCGTGCAGGCGCTCGAGGCGATGCTGTTCGCGCTGGACGCGGCGCGGGCCCTCGCCCCGCCCGCCGTCAGCCTCGGCGCACACGTCCTAGACGACTGCGACAACGACACCTATGGATTAGAAATGGCGCTCGACTTCATCAAAGGTTCCATCAGCAACATCGACGACGCGGAGTACGCTTGCAACGCGAGTGCCGTGCGGAAGGTCATCTCCGGAGTGGTGGGCGCGGCTTCCAGTGTCACGTCCGTGCAAGTCGCTAACCTGTTGCGACTGTTCAAGATACCGCAGGTATCATTTTTTTCGACTTCTCCAGAGCTTTCGAACAAAGCTCGATTTGAATACTTTACTAGAACTATTCCATCTGACCTTCACCAAGTGCGGGCTATGGTAGAGATAGTCAAGAAATTGGGCTGGCGCTATGTTTCTATCATTTACGAAGAATCCAACTACGGCATTAAGGCTTTTGAAGAACTGGAAACTTTACTGGCACGCAATGAGATCTGTATCGCGGTGAAGGAGAGGCTGGTAAAAGATTCGGGGGAGGCGGACGAGCGAGCTTACGACAACATCGTGGCGCGTCTCCTGTCGCGACCGCGGGCCCGAGGAGTGATCGTGTTCGGCTCGGATCAAGAAGTGGCAGGCGTGATGGCGGCGGTGGCTCGCGCGGGGGCCACCGGAGCCTTCAGCTGGGTCGGCTCGGACGGCTGGAGCGCTCGGGCGCTGGTCTCGGACGGAAACGAGGAGGCCGTCGAGGGAACTATCAGCGTCCAGCCCCAGGCTAACCCTGTGCGCGGCTTCAAGGAGTACTTCCTCAACCTCACCGTAGAGAACAACCCGAGGAACCCCTGGTTCGTAGAGTTCTGGGAGGATCAGTTCCGGTGTCGATACCCGGGCAGCGCCCGCACGCCCTACAACGCTCAGTACTCGCGGCTGTGCTCGGGGCGGGAGCGACTCTCCGCAGGGAACATAGAGTTCGAGGCTCAGCTGCAGTTCGTCACCGACGCCGTGCTTGCGTTCGCGCACGCCATTAGAGACATGCACGCGGACCTGTGCGGAGGAGAGCCGGGACTGTGCGAGGCGATGCGACCCGCCAGCGGACCGACGCTGTTGCGCTACCTGCGACAAGTTCGCTTCACAGGTCTGAGCGGGGATGAGTTCCACTTCGACAGCAACGGCGACGGCCCCGCGCGATACAACATCCTGCACTTCAAGCAGCTGTCGCCCGGGGTCTACCGCTGGCTGAAGGTCGGCCGCTACCTCGACGGAGAGCTCGAACTGGACATCGACGATATCCAGTTCAAGTGGGAATCAGCCCGACCCCCGGAGTCGGTGTGCAGCGTGGAGTGCGAGCTCGGACAAGCCAAGCAGTATGTGGAGGGTGAGAGCTGCTGCTGGCATTGCTTCAACTGCACGCAGTACGAGGTCCGGTCGCCGAGCTCGGAGACGGCGTGCGTGGAGTGCGCGCTGGGCACGCTGCCGGACCCGCGCCGGATGCGGTGCGCGCCCGTGCCGGAGCTCTATCTGCGACCCGACTCTGCTGCCGCTGTCGCCGCTATGACTTTCTCTTCCCTTGGAATCATCCTCACAGTGTTCGTGGTGTGCGTGTGGGCGGCGCGCGGCTCGACGCCGGTGGTGCGCGCCAGCGGCCGGGAGCTGTCGCTGGTGCTGCTCGCGGGGATCCTCATGTGCTACCTCGTCACCTTTGCACTCGTCTTCAGACCAACAGATGTTCTCTGCGCTCTTCAGCGTTTCGGCACCGGGTTCTGCTTCACGGTGGTGTATGCGGCGCTGCTCACAAAGACCAACCGCATCGCTCGTATCTTCGCAGCCAGCAAGCACTCCGCGCGCCGACCCTCGCTCATCTCGCCCAAGTCGCAGCTGCTCATCTGTTCTGCCCTCGTATCTGTCCAGGTGGTGATAGTGGTGGTGTGGCAGATAGTGTCACCGGCGCGCGCCATACATCACTACCCGACGCGCGAAGACAACATGCTGGTCTGCGACTCATACGTGGATGCGTCCTACACCATCGCCTTCTTCTACCCCGTGGTGCTCATCCTGGTGTGTACAGTATACGCCGTTCTGACGCGCAAGATACCAGAAGCCTTCAACGAAAGCAAACACATAGGGTTCACCATGTACACGACGTGCGTGATCTGGCTGGCGTTCGTGCCGCTGTACTTCGGGACGGCGAGCCACGTGCCCCTGCGCGTCACCAGCATGGCCATCACGATATCGCTCAGCGCCAGCGTCACGCTCGTCTGCCTGTTCTCGCCGAAGGTGTATATAATCCTGGTGCGTCCGGAACGAAACGTGCGTGCAAGCCTGATGCCGACGCGATGGAGGGGAGGGGTGAGGGGCGGCGCGGCGGGAGGGGCGGTGTGCGCGGCGCTGGTCGGCACCGCTCCCCTCCCTCGCGCACCCGACCTTACACCCTCCACCGACCTTTCCACCTTGAACGAACCTGAGCGCTCGTCAACGGATCGTCAAGTGCAGACGGAGGAGGAGTCGGAGCTGTACCCGCGTCTGCGTCGCGCGGTGGAAGAGAACGGCGTTCGGCTGGACGCGCGGCAATTAGAGGCGCTGCGGGCCGGGCTGCCGGCGTTCGGTGCTGGAGGTTCGGCGCCGCCCTCGGCCGAGCTGGCCGTGGCGGTGCTCGGCGCGGACCTGCCCTTATAG
Protein
MSAAARVLLATLLATAALASHADRTHLANISSALTNPTTQLFYDDLLPSHILRSKRNAHTLIATFDISKIFKRKKLQSKQGDSPNPTPSSLLSDNLSASMVAELLATSLANVSLVVDTTPLTDPIELTTSLVSNSTISTKRVSTPFTQISNPKNSLKLKSKSESKGVDASVSSRVGVKKKVWENETVWPVKHAAVVDGDIILGGLMMVHRRSEVATCGPVMAQGGVQALEAMLFALDAARALAPPAVSLGAHVLDDCDNDTYGLEMALDFIKGSISNIDDAEYACNASAVRKVISGVVGAASSVTSVQVANLLRLFKIPQVSFFSTSPELSNKARFEYFTRTIPSDLHQVRAMVEIVKKLGWRYVSIIYEESNYGIKAFEELETLLARNEICIAVKERLVKDSGEADERAYDNIVARLLSRPRARGVIVFGSDQEVAGVMAAVARAGATGAFSWVGSDGWSARALVSDGNEEAVEGTISVQPQANPVRGFKEYFLNLTVENNPRNPWFVEFWEDQFRCRYPGSARTPYNAQYSRLCSGRERLSAGNIEFEAQLQFVTDAVLAFAHAIRDMHADLCGGEPGLCEAMRPASGPTLLRYLRQVRFTGLSGDEFHFDSNGDGPARYNILHFKQLSPGVYRWLKVGRYLDGELELDIDDIQFKWESARPPESVCSVECELGQAKQYVEGESCCWHCFNCTQYEVRSPSSETACVECALGTLPDPRRMRCAPVPELYLRPDSAAAVAAMTFSSLGIILTVFVVCVWAARGSTPVVRASGRELSLVLLAGILMCYLVTFALVFRPTDVLCALQRFGTGFCFTVVYAALLTKTNRIARIFAASKHSARRPSLISPKSQLLICSALVSVQVVIVVVWQIVSPARAIHHYPTREDNMLVCDSYVDASYTIAFFYPVVLILVCTVYAVLTRKIPEAFNESKHIGFTMYTTCVIWLAFVPLYFGTASHVPLRVTSMAITISLSASVTLVCLFSPKVYIILVRPERNVRASLMPTRWRGGVRGGAAGGAVCAALVGTAPLPRAPDLTPSTDLSTLNEPERSSTDRQVQTEEESELYPRLRRAVEENGVRLDARQLEALRAGLPAFGAGGSAPPSAELAVAVLGADLPL
Summary
Similarity
Belongs to the G-protein coupled receptor 3 family.
Uniprot
A0A2W1BGB3
A0A2A4JI84
A0A212FEA2
A0A194RGF0
A0A2W1BFP1
A0A194Q8J6
+ More
A0A0P9ACD0 A0A1I8M0J9 A0A0J9TWB7 A0A0R3NL13 A0A0R1DM99 B4KP51 A0A0K8U462 B3MFX4 A1Z7F4 B4J5X5 A0A0Q5WKX7 A0A0B4KF15 B4LLC1 B7YZT0 Q70GQ8 A0A0J9R8A8 B4P2U6 A0A3B0IZT4 A1Z7F5 B3N8N5 B4GDF5 B4HRY5 A0A1W4UJR8 A0A154PQC1 B5DZA5 K7J0X5 A0A088AQD5 Q75QW6 A0A195B3M6 B4MQ18 A0A195DLK5 A0A1B0C9E3 A0A151WWG1 A0A195D7C2 W8BYX6 A0A0L7LCG4 A0A195FTU1 A0A1J1J939 A0A0K8VB57 A0A182NLU4 A0A182RUV7 A0A182YJ34 A0A182PRF3 A0A084WKZ5 Q7Q9V1 T1I399 A0A1W7R6Q5 A0A182X0Z4 A0A2S2PI57 A0A2H8TP95 A0A182HPF8 Q16UR9 A0A1J0FAV9 A0A1J0FAU2 A0A0P5MRK9 A0A0P5PC66 A0A0P5MUK6 A0A0P5N2A9 A0A0P6H970 A0A0P5V439 A0A0P5Z5H7 A0A0P5MY78 A0A0P5M842 A0A0P5A5G2 A0A0P5Y8F6 A0A0P5NTS2 A0A0P5AC45 A0A0N8BRE4 A0A0P5N2P5 A0A0P5N7E1 A0A0N8D1A2 A0A0P5W0X8 A0A0P6DRG4 A0A0P5NBT2 A0A0P5ANY0 A0A0P5MPZ1 A0A0P5MT02 A0A0N8BX12 A0A0P5M855 A0A0P5NI69 A0A0P5Q8Y9 A0A0P5MQ28 A0A0P5N9Q5 A0A0P5QD41 A0A0P5NWY8 A0A0P5V9J3 A0A0P5AF09 A0A0P5A955 A0A0N8BQE4
A0A0P9ACD0 A0A1I8M0J9 A0A0J9TWB7 A0A0R3NL13 A0A0R1DM99 B4KP51 A0A0K8U462 B3MFX4 A1Z7F4 B4J5X5 A0A0Q5WKX7 A0A0B4KF15 B4LLC1 B7YZT0 Q70GQ8 A0A0J9R8A8 B4P2U6 A0A3B0IZT4 A1Z7F5 B3N8N5 B4GDF5 B4HRY5 A0A1W4UJR8 A0A154PQC1 B5DZA5 K7J0X5 A0A088AQD5 Q75QW6 A0A195B3M6 B4MQ18 A0A195DLK5 A0A1B0C9E3 A0A151WWG1 A0A195D7C2 W8BYX6 A0A0L7LCG4 A0A195FTU1 A0A1J1J939 A0A0K8VB57 A0A182NLU4 A0A182RUV7 A0A182YJ34 A0A182PRF3 A0A084WKZ5 Q7Q9V1 T1I399 A0A1W7R6Q5 A0A182X0Z4 A0A2S2PI57 A0A2H8TP95 A0A182HPF8 Q16UR9 A0A1J0FAV9 A0A1J0FAU2 A0A0P5MRK9 A0A0P5PC66 A0A0P5MUK6 A0A0P5N2A9 A0A0P6H970 A0A0P5V439 A0A0P5Z5H7 A0A0P5MY78 A0A0P5M842 A0A0P5A5G2 A0A0P5Y8F6 A0A0P5NTS2 A0A0P5AC45 A0A0N8BRE4 A0A0P5N2P5 A0A0P5N7E1 A0A0N8D1A2 A0A0P5W0X8 A0A0P6DRG4 A0A0P5NBT2 A0A0P5ANY0 A0A0P5MPZ1 A0A0P5MT02 A0A0N8BX12 A0A0P5M855 A0A0P5NI69 A0A0P5Q8Y9 A0A0P5MQ28 A0A0P5N9Q5 A0A0P5QD41 A0A0P5NWY8 A0A0P5V9J3 A0A0P5AF09 A0A0P5A955 A0A0N8BQE4
Pubmed
EMBL
KZ150487
PZC70703.1
NWSH01001473
PCG71140.1
AGBW02008952
OWR52095.1
+ More
KQ460211 KPJ16524.1 KZ150080 PZC73882.1 KQ459302 KPJ01848.1 CH902619 KPU75778.1 CM002911 KMY92273.1 KMY92274.1 KMY92275.1 CM000071 KRT01640.1 CM000157 KRJ98364.1 CH933808 EDW10117.2 GDHF01030797 JAI21517.1 EDV35656.2 AE013599 AAF59081.3 CH916367 EDW00818.1 CH954177 KQS70875.1 AGB93339.1 CH940648 EDW60858.2 ACL83073.1 AJ581534 CAE46392.1 KMY92276.1 EDW89357.2 OUUW01000001 SPP73687.1 AAF59082.3 EDV59512.1 CH479181 EDW31631.1 CH480816 EDW46948.1 KQ435036 KZC14082.1 EDY68791.2 AB161182 BAD08344.1 KQ976641 KYM78799.1 CH963849 EDW74207.1 KQ980765 KYN13374.1 AJWK01002248 KQ982691 KYQ52223.1 KQ976749 KYN08785.1 GAMC01002043 JAC04513.1 JTDY01001683 KOB73168.1 KQ981272 KYN43846.1 CVRI01000074 CRL08038.1 GDHF01016218 JAI36096.1 ATLV01024163 KE525350 KFB50889.1 AAAB01008900 EAA09380.4 ACPB03010517 GEHC01000817 JAV46828.1 GGMR01016299 MBY28918.1 GFXV01003936 MBW15741.1 APCN01002995 CH477615 EAT38269.2 KU986890 APC26119.1 KU986887 APC26116.1 GDIQ01152312 JAK99413.1 GDIQ01152309 JAK99416.1 GDIQ01155159 JAK96566.1 GDIQ01152306 JAK99419.1 GDIQ01044510 GDIQ01044509 JAN50227.1 GDIP01105826 JAL97888.1 GDIP01206565 GDIP01061385 JAM42330.1 GDIQ01160960 GDIQ01160959 JAK90766.1 GDIQ01159361 JAK92364.1 GDIP01208214 GDIP01070778 JAJ15188.1 GDIP01110233 GDIP01061387 JAM42328.1 GDIQ01159358 JAK92367.1 GDIP01201160 JAJ22242.1 GDIQ01152307 JAK99418.1 GDIQ01159359 GDIQ01157464 JAK92366.1 GDIQ01157466 JAK94259.1 GDIP01078631 JAM25084.1 GDIP01093581 JAM10134.1 GDIQ01073442 JAN21295.1 GDIQ01155162 JAK96563.1 GDIP01199863 JAJ23539.1 GDIQ01155160 JAK96565.1 GDIQ01152311 JAK99414.1 GDIQ01136563 JAL15163.1 GDIQ01159360 JAK92365.1 GDIQ01152310 JAK99415.1 GDIQ01139070 JAL12656.1 GDIQ01155105 JAK96620.1 GDIQ01152313 JAK99412.1 GDIQ01122016 JAL29710.1 GDIQ01136567 JAL15159.1 GDIP01103029 GDIP01103028 JAM00686.1 GDIP01199864 JAJ23538.1 GDIP01202362 JAJ21040.1 GDIQ01155107 JAK96618.1
KQ460211 KPJ16524.1 KZ150080 PZC73882.1 KQ459302 KPJ01848.1 CH902619 KPU75778.1 CM002911 KMY92273.1 KMY92274.1 KMY92275.1 CM000071 KRT01640.1 CM000157 KRJ98364.1 CH933808 EDW10117.2 GDHF01030797 JAI21517.1 EDV35656.2 AE013599 AAF59081.3 CH916367 EDW00818.1 CH954177 KQS70875.1 AGB93339.1 CH940648 EDW60858.2 ACL83073.1 AJ581534 CAE46392.1 KMY92276.1 EDW89357.2 OUUW01000001 SPP73687.1 AAF59082.3 EDV59512.1 CH479181 EDW31631.1 CH480816 EDW46948.1 KQ435036 KZC14082.1 EDY68791.2 AB161182 BAD08344.1 KQ976641 KYM78799.1 CH963849 EDW74207.1 KQ980765 KYN13374.1 AJWK01002248 KQ982691 KYQ52223.1 KQ976749 KYN08785.1 GAMC01002043 JAC04513.1 JTDY01001683 KOB73168.1 KQ981272 KYN43846.1 CVRI01000074 CRL08038.1 GDHF01016218 JAI36096.1 ATLV01024163 KE525350 KFB50889.1 AAAB01008900 EAA09380.4 ACPB03010517 GEHC01000817 JAV46828.1 GGMR01016299 MBY28918.1 GFXV01003936 MBW15741.1 APCN01002995 CH477615 EAT38269.2 KU986890 APC26119.1 KU986887 APC26116.1 GDIQ01152312 JAK99413.1 GDIQ01152309 JAK99416.1 GDIQ01155159 JAK96566.1 GDIQ01152306 JAK99419.1 GDIQ01044510 GDIQ01044509 JAN50227.1 GDIP01105826 JAL97888.1 GDIP01206565 GDIP01061385 JAM42330.1 GDIQ01160960 GDIQ01160959 JAK90766.1 GDIQ01159361 JAK92364.1 GDIP01208214 GDIP01070778 JAJ15188.1 GDIP01110233 GDIP01061387 JAM42328.1 GDIQ01159358 JAK92367.1 GDIP01201160 JAJ22242.1 GDIQ01152307 JAK99418.1 GDIQ01159359 GDIQ01157464 JAK92366.1 GDIQ01157466 JAK94259.1 GDIP01078631 JAM25084.1 GDIP01093581 JAM10134.1 GDIQ01073442 JAN21295.1 GDIQ01155162 JAK96563.1 GDIP01199863 JAJ23539.1 GDIQ01155160 JAK96565.1 GDIQ01152311 JAK99414.1 GDIQ01136563 JAL15163.1 GDIQ01159360 JAK92365.1 GDIQ01152310 JAK99415.1 GDIQ01139070 JAL12656.1 GDIQ01155105 JAK96620.1 GDIQ01152313 JAK99412.1 GDIQ01122016 JAL29710.1 GDIQ01136567 JAL15159.1 GDIP01103029 GDIP01103028 JAM00686.1 GDIP01199864 JAJ23538.1 GDIP01202362 JAJ21040.1 GDIQ01155107 JAK96618.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000007801
UP000095301
+ More
UP000001819 UP000002282 UP000009192 UP000000803 UP000001070 UP000008711 UP000008792 UP000268350 UP000008744 UP000001292 UP000192221 UP000076502 UP000002358 UP000005203 UP000078540 UP000007798 UP000078492 UP000092461 UP000075809 UP000078542 UP000037510 UP000078541 UP000183832 UP000075884 UP000075900 UP000076408 UP000075885 UP000030765 UP000007062 UP000015103 UP000076407 UP000075840 UP000008820
UP000001819 UP000002282 UP000009192 UP000000803 UP000001070 UP000008711 UP000008792 UP000268350 UP000008744 UP000001292 UP000192221 UP000076502 UP000002358 UP000005203 UP000078540 UP000007798 UP000078492 UP000092461 UP000075809 UP000078542 UP000037510 UP000078541 UP000183832 UP000075884 UP000075900 UP000076408 UP000075885 UP000030765 UP000007062 UP000015103 UP000076407 UP000075840 UP000008820
Pfam
Interpro
IPR038550
GPCR_3_9-Cys_sf
+ More
IPR017978 GPCR_3_C
IPR000162 GPCR_3_mtglu_rcpt
IPR001828 ANF_lig-bd_rcpt
IPR000337 GPCR_3
IPR011500 GPCR_3_9-Cys_dom
IPR028082 Peripla_BP_I
IPR017979 GPCR_3_CS
IPR000330 SNF2_N
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR014001 Helicase_ATP-bd
IPR020838 DBINO
IPR001650 Helicase_C
IPR017978 GPCR_3_C
IPR000162 GPCR_3_mtglu_rcpt
IPR001828 ANF_lig-bd_rcpt
IPR000337 GPCR_3
IPR011500 GPCR_3_9-Cys_dom
IPR028082 Peripla_BP_I
IPR017979 GPCR_3_CS
IPR000330 SNF2_N
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR014001 Helicase_ATP-bd
IPR020838 DBINO
IPR001650 Helicase_C
Gene 3D
CDD
ProteinModelPortal
A0A2W1BGB3
A0A2A4JI84
A0A212FEA2
A0A194RGF0
A0A2W1BFP1
A0A194Q8J6
+ More
A0A0P9ACD0 A0A1I8M0J9 A0A0J9TWB7 A0A0R3NL13 A0A0R1DM99 B4KP51 A0A0K8U462 B3MFX4 A1Z7F4 B4J5X5 A0A0Q5WKX7 A0A0B4KF15 B4LLC1 B7YZT0 Q70GQ8 A0A0J9R8A8 B4P2U6 A0A3B0IZT4 A1Z7F5 B3N8N5 B4GDF5 B4HRY5 A0A1W4UJR8 A0A154PQC1 B5DZA5 K7J0X5 A0A088AQD5 Q75QW6 A0A195B3M6 B4MQ18 A0A195DLK5 A0A1B0C9E3 A0A151WWG1 A0A195D7C2 W8BYX6 A0A0L7LCG4 A0A195FTU1 A0A1J1J939 A0A0K8VB57 A0A182NLU4 A0A182RUV7 A0A182YJ34 A0A182PRF3 A0A084WKZ5 Q7Q9V1 T1I399 A0A1W7R6Q5 A0A182X0Z4 A0A2S2PI57 A0A2H8TP95 A0A182HPF8 Q16UR9 A0A1J0FAV9 A0A1J0FAU2 A0A0P5MRK9 A0A0P5PC66 A0A0P5MUK6 A0A0P5N2A9 A0A0P6H970 A0A0P5V439 A0A0P5Z5H7 A0A0P5MY78 A0A0P5M842 A0A0P5A5G2 A0A0P5Y8F6 A0A0P5NTS2 A0A0P5AC45 A0A0N8BRE4 A0A0P5N2P5 A0A0P5N7E1 A0A0N8D1A2 A0A0P5W0X8 A0A0P6DRG4 A0A0P5NBT2 A0A0P5ANY0 A0A0P5MPZ1 A0A0P5MT02 A0A0N8BX12 A0A0P5M855 A0A0P5NI69 A0A0P5Q8Y9 A0A0P5MQ28 A0A0P5N9Q5 A0A0P5QD41 A0A0P5NWY8 A0A0P5V9J3 A0A0P5AF09 A0A0P5A955 A0A0N8BQE4
A0A0P9ACD0 A0A1I8M0J9 A0A0J9TWB7 A0A0R3NL13 A0A0R1DM99 B4KP51 A0A0K8U462 B3MFX4 A1Z7F4 B4J5X5 A0A0Q5WKX7 A0A0B4KF15 B4LLC1 B7YZT0 Q70GQ8 A0A0J9R8A8 B4P2U6 A0A3B0IZT4 A1Z7F5 B3N8N5 B4GDF5 B4HRY5 A0A1W4UJR8 A0A154PQC1 B5DZA5 K7J0X5 A0A088AQD5 Q75QW6 A0A195B3M6 B4MQ18 A0A195DLK5 A0A1B0C9E3 A0A151WWG1 A0A195D7C2 W8BYX6 A0A0L7LCG4 A0A195FTU1 A0A1J1J939 A0A0K8VB57 A0A182NLU4 A0A182RUV7 A0A182YJ34 A0A182PRF3 A0A084WKZ5 Q7Q9V1 T1I399 A0A1W7R6Q5 A0A182X0Z4 A0A2S2PI57 A0A2H8TP95 A0A182HPF8 Q16UR9 A0A1J0FAV9 A0A1J0FAU2 A0A0P5MRK9 A0A0P5PC66 A0A0P5MUK6 A0A0P5N2A9 A0A0P6H970 A0A0P5V439 A0A0P5Z5H7 A0A0P5MY78 A0A0P5M842 A0A0P5A5G2 A0A0P5Y8F6 A0A0P5NTS2 A0A0P5AC45 A0A0N8BRE4 A0A0P5N2P5 A0A0P5N7E1 A0A0N8D1A2 A0A0P5W0X8 A0A0P6DRG4 A0A0P5NBT2 A0A0P5ANY0 A0A0P5MPZ1 A0A0P5MT02 A0A0N8BX12 A0A0P5M855 A0A0P5NI69 A0A0P5Q8Y9 A0A0P5MQ28 A0A0P5N9Q5 A0A0P5QD41 A0A0P5NWY8 A0A0P5V9J3 A0A0P5AF09 A0A0P5A955 A0A0N8BQE4
PDB
6N52
E-value=5.53488e-150,
Score=1366
Ontologies
GO
Topology
Subcellular location
Cell membrane
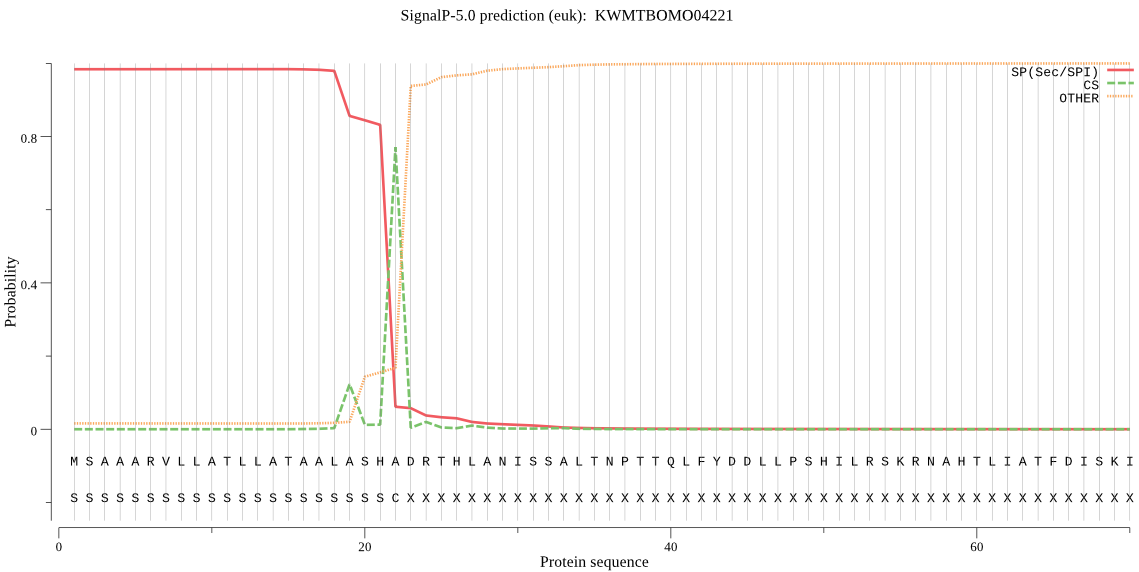
SignalP
Position: 1 - 22,
Likelihood: 0.983697
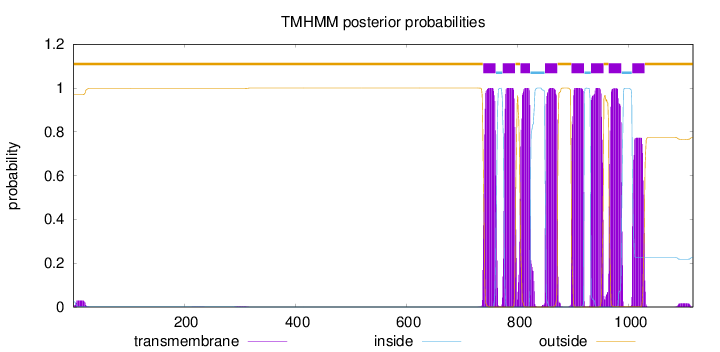
Length:
1115
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
170.84575
Exp number, first 60 AAs:
0.53213
Total prob of N-in:
0.03054
outside
1 - 737
TMhelix
738 - 760
inside
761 - 772
TMhelix
773 - 795
outside
796 - 804
TMhelix
805 - 822
inside
823 - 848
TMhelix
849 - 871
outside
872 - 896
TMhelix
897 - 919
inside
920 - 931
TMhelix
932 - 954
outside
955 - 963
TMhelix
964 - 986
inside
987 - 1005
TMhelix
1006 - 1028
outside
1029 - 1115
Population Genetic Test Statistics
Pi
225.741754
Theta
204.347278
Tajima's D
0.599075
CLR
0.032321
CSRT
0.538223088845558
Interpretation
Uncertain
