Gene
KWMTBOMO04202
Pre Gene Modal
BGIBMGA008647
Annotation
hypothetical_protein_KGM_19575_[Danaus_plexippus]
Full name
Dystrophin, isoforms A/C/F/G/H
+ More
Dystrophin, isoform B
Dystrophin, isoform B
Alternative Name
Protein detached
Location in the cell
Nuclear Reliability : 2.742
Sequence
CDS
ATGAGTAAGCTGGGAGAGCTTAAGGATCTGCAATCGGAGATCGAGACACACAGGAATGTGTACGCATCGCTCACTGGAACTGGCCGGCGTCTCCTGGGCAGCCTGTCTTCTCAGGAAGACGCTGTCATGCTGCAGAGGAGACTGGATGAGATGAATCAGAGATGGCATCACTTGAAAGCGAAGAGTATGGCTATCAGAAACCGCCTTGAGAGCAACGCCGAACATTGGTCCGCATTGCTGCTGTCTCTCCGGGAGTTAACTGAGTGGGTCATCAGGAAAGAGACAGAATTGAACGCTCTAGCACCTCCGAGGGGAGACCTCACCGCCCTCCTAAAGCAGCAAGATGATCATCGCGCCTTCAGGAGACAATTGGAAGACAAGCGTCCAGTGGTGGAGAGCAACCTCCTCAGCGGAAGGCAGTACATCGCCAACGAACCACCTCTCTCCGACACCAGTGATACTGAACCATCTCGGGACAGCGAAGGCGACTCCCGCGGCTACCGGTCTGCCGAGGAACAGGCCCGCGAGCTGGCCCGCTCTATCCGCCGCGAGGTGGCCAAGCTCGCTGACAAGTGGAACAACCTGGTCGACCGCAGCGACGCGTGGGGCAGGTGCTTGGATGATGCTGTACAGGTATGTCTAGAACTTAAGCTTACCTTTATTAATAAAACTAGCGGACCGTTCCGGCTCCGCTCGGGTCTTTAA
Protein
MSKLGELKDLQSEIETHRNVYASLTGTGRRLLGSLSSQEDAVMLQRRLDEMNQRWHHLKAKSMAIRNRLESNAEHWSALLLSLRELTEWVIRKETELNALAPPRGDLTALLKQQDDHRAFRRQLEDKRPVVESNLLSGRQYIANEPPLSDTSDTEPSRDSEGDSRGYRSAEEQARELARSIRREVAKLADKWNNLVDRSDAWGRCLDDAVQVCLELKLTFINKTSGPFRLRSGL
Summary
Description
Required for the maintenance of appropriate synaptic retrograde communication and the stabilization of muscle cell architecture or physiology. Both det and Dg are required for maintenance of early dpp signaling in the presumptive crossvein. Isoform A is not required to maintain muscle integrity, but plays a role in neuromuscular homeostasis by regulating neurotransmitter release. May play a role in anchoring the cytoskeleton to the plasma membrane.
Required for the maintenance of appropriate synaptic retrograde communication and the stabilization of muscle cell architecture or physiology. May play a role in anchoring the cytoskeleton to the plasma membrane.
Required for the maintenance of appropriate synaptic retrograde communication and the stabilization of muscle cell architecture or physiology. May play a role in anchoring the cytoskeleton to the plasma membrane.
Subunit
Component of the dystrophin associated protein complex (DAPC). Interacts with Dg, via the Dg WW domain binding sites.
Keywords
Actin-binding
Alternative splicing
Calcium
Cell membrane
Complete proteome
Cytoplasm
Cytoskeleton
Membrane
Metal-binding
Phosphoprotein
Reference proteome
Repeat
Zinc
Zinc-finger
Feature
chain Dystrophin, isoforms A/C/F/G/H
splice variant In isoform F.
splice variant In isoform F.
Uniprot
A0A212EQW8
A0A194PWC9
A0A1E1WJ00
A0A194RDD7
A0A1B6EW19
A0A1B6KWY7
+ More
A0A1B6LPQ0 A0A088AJ44 A0A1B6I1K0 A0A1B6IKG4 A0A310STA5 A0A232FMZ1 A0A1B6LEA1 A0A1B6KJ06 A0A158NE51 V9ICT2 A0A2A3EC27 E0VJZ9 A0A1B6HMA6 A0A0C9RVM7 A0A0L7R7U4 A0A0N0BDW7 A0A1B0G180 A0A336MUW4 A0A1A9X1L7 A0A1A9XP77 A0A336MMA3 A0A0K8TBX1 A0A0K8T9U8 A0A0K8T9F1 A0A0K8T9F7 A0A0K8TAJ4 A0A0A9YSD3 A0A0A9ZD72 A0A0A9YQV7 A0A0A9ZD78 A0A2J7PV18 A0A0A9Z5J2 A0A0A9Z8S5 A0A0K8T9D3 A0A0A9Z3C7 A0A0A9Z8T0 A0A0A9Z5J6 A0A0A9Z770 A0A0A9YLR0 A0A0A9Z766 A0A2J7PV24 A0A2J7PV58 A0A3L8DXZ2 A0A069DY06 A0A1L8E575 A0A224X947 A0A023F0P7 A0A0Q9WZ85 B4KCK7 B4GLW3 A0A0Q9X7F0 A0A0Q9X128 A0A0Q9WZ57 A0A0Q9X5Z0 A0A0Q9XB81 A0A0Q9WZP4 A0A0Q9X9Z4 A0A0Q9XBK8 A0A0Q9WZ87 A0A0B4KGB9 B4JYZ5 B4LZ03 A0A0Q9WGJ9 A0A0Q9WHH3 A0A0Q9WGH2 A0A0Q9WRQ6 A0A0Q9WSC2 A0A0Q9WGP2 A0A0B4KHR4 A0A0Q9WNZ9 A0A1W4W9F8 A0A0Q9WQI2 A0A0Q9WSU5 A0A0Q9WH19 Q9VDW6-2 A0A0B4KGH0 A0A0R3NKF7 A0A0Q9WU35 A0A0Q9WU41 A0A0R3NR30 Q9VDW3 A0A0Q9WIB5 A0A0Q9WGQ7 A0A0R3NK00 A0A0M4EJD4 A0A0R3NKA5 A0A0Q9WSF8 A0A0R3NPN5 Q9VDW6 A0A1W4W848 Q293M7
A0A1B6LPQ0 A0A088AJ44 A0A1B6I1K0 A0A1B6IKG4 A0A310STA5 A0A232FMZ1 A0A1B6LEA1 A0A1B6KJ06 A0A158NE51 V9ICT2 A0A2A3EC27 E0VJZ9 A0A1B6HMA6 A0A0C9RVM7 A0A0L7R7U4 A0A0N0BDW7 A0A1B0G180 A0A336MUW4 A0A1A9X1L7 A0A1A9XP77 A0A336MMA3 A0A0K8TBX1 A0A0K8T9U8 A0A0K8T9F1 A0A0K8T9F7 A0A0K8TAJ4 A0A0A9YSD3 A0A0A9ZD72 A0A0A9YQV7 A0A0A9ZD78 A0A2J7PV18 A0A0A9Z5J2 A0A0A9Z8S5 A0A0K8T9D3 A0A0A9Z3C7 A0A0A9Z8T0 A0A0A9Z5J6 A0A0A9Z770 A0A0A9YLR0 A0A0A9Z766 A0A2J7PV24 A0A2J7PV58 A0A3L8DXZ2 A0A069DY06 A0A1L8E575 A0A224X947 A0A023F0P7 A0A0Q9WZ85 B4KCK7 B4GLW3 A0A0Q9X7F0 A0A0Q9X128 A0A0Q9WZ57 A0A0Q9X5Z0 A0A0Q9XB81 A0A0Q9WZP4 A0A0Q9X9Z4 A0A0Q9XBK8 A0A0Q9WZ87 A0A0B4KGB9 B4JYZ5 B4LZ03 A0A0Q9WGJ9 A0A0Q9WHH3 A0A0Q9WGH2 A0A0Q9WRQ6 A0A0Q9WSC2 A0A0Q9WGP2 A0A0B4KHR4 A0A0Q9WNZ9 A0A1W4W9F8 A0A0Q9WQI2 A0A0Q9WSU5 A0A0Q9WH19 Q9VDW6-2 A0A0B4KGH0 A0A0R3NKF7 A0A0Q9WU35 A0A0Q9WU41 A0A0R3NR30 Q9VDW3 A0A0Q9WIB5 A0A0Q9WGQ7 A0A0R3NK00 A0A0M4EJD4 A0A0R3NKA5 A0A0Q9WSF8 A0A0R3NPN5 Q9VDW6 A0A1W4W848 Q293M7
Pubmed
EMBL
AGBW02013189
OWR43896.1
KQ459589
KPI97632.1
GDQN01004187
JAT86867.1
+ More
KQ460367 KPJ15479.1 GECZ01027555 JAS42214.1 GEBQ01024019 JAT15958.1 GEBQ01014295 JAT25682.1 GECU01026907 JAS80799.1 GECU01020297 JAS87409.1 KQ760629 OAD59723.1 NNAY01000019 OXU31889.1 GEBQ01018143 JAT21834.1 GEBQ01028823 JAT11154.1 ADTU01012837 ADTU01012838 ADTU01012839 ADTU01012840 ADTU01012841 ADTU01012842 ADTU01012843 ADTU01012844 ADTU01012845 ADTU01012846 JR038061 AEY58099.1 KZ288290 PBC29240.1 DS235235 EEB13705.1 GECU01031869 JAS75837.1 GBYB01011751 JAG81518.1 KQ414637 KOC66952.1 KQ435851 KOX70714.1 CCAG010022463 UFQT01002783 SSX34056.1 UFQT01001358 SSX30199.1 GBRD01002769 JAG63052.1 GBRD01003655 JAG62166.1 GBRD01003657 JAG62164.1 GBRD01003638 JAG62183.1 GBRD01003651 JAG62170.1 GBHO01009596 JAG34008.1 GBHO01003854 JAG39750.1 GBHO01009598 JAG34006.1 GBHO01003849 JAG39755.1 NEVH01020989 PNF20188.1 GBHO01003850 JAG39754.1 GBHO01003851 JAG39753.1 GBRD01003659 JAG62162.1 GBHO01003847 JAG39757.1 GBHO01003846 JAG39758.1 GBHO01003845 JAG39759.1 GBHO01003848 JAG39756.1 GBHO01009597 JAG34007.1 GBHO01003853 JAG39751.1 PNF20189.1 PNF20190.1 QOIP01000002 RLU25330.1 GBGD01000006 JAC88883.1 GFDF01000226 JAV13858.1 GFTR01009002 JAW07424.1 GBBI01003645 JAC15067.1 CH933806 KRG01228.1 EDW14826.1 CH479185 EDW38537.1 KRG01220.1 KRG01221.1 KRG01225.1 KRG01222.1 KRG01226.1 KRG01224.1 KRG01229.1 KRG01223.1 KRG01219.1 KRG01227.1 AE014297 AGB96113.1 CH916378 EDV98610.1 CH940650 EDW68106.1 KRF83673.1 KRF83657.1 KRF83660.1 KRF83666.1 KRF83661.1 KRF83663.1 KRF83668.1 KRF83656.1 AGB96110.1 KRF83671.1 KRF83665.1 KRF83659.1 KRF83667.1 AF277386 AF297644 AF300456 AF302236 AAF55673.2 AGB96112.1 CM000070 KRT01348.1 KRF83669.1 KRF83664.1 KRT01360.1 AF300294 X99757 AAF55676.3 AAK15257.1 KRF83670.1 KRF83672.1 KRT01345.1 CP012526 ALC46996.1 KRT01342.1 KRF83662.1 KRT01343.1 EAL29187.4
KQ460367 KPJ15479.1 GECZ01027555 JAS42214.1 GEBQ01024019 JAT15958.1 GEBQ01014295 JAT25682.1 GECU01026907 JAS80799.1 GECU01020297 JAS87409.1 KQ760629 OAD59723.1 NNAY01000019 OXU31889.1 GEBQ01018143 JAT21834.1 GEBQ01028823 JAT11154.1 ADTU01012837 ADTU01012838 ADTU01012839 ADTU01012840 ADTU01012841 ADTU01012842 ADTU01012843 ADTU01012844 ADTU01012845 ADTU01012846 JR038061 AEY58099.1 KZ288290 PBC29240.1 DS235235 EEB13705.1 GECU01031869 JAS75837.1 GBYB01011751 JAG81518.1 KQ414637 KOC66952.1 KQ435851 KOX70714.1 CCAG010022463 UFQT01002783 SSX34056.1 UFQT01001358 SSX30199.1 GBRD01002769 JAG63052.1 GBRD01003655 JAG62166.1 GBRD01003657 JAG62164.1 GBRD01003638 JAG62183.1 GBRD01003651 JAG62170.1 GBHO01009596 JAG34008.1 GBHO01003854 JAG39750.1 GBHO01009598 JAG34006.1 GBHO01003849 JAG39755.1 NEVH01020989 PNF20188.1 GBHO01003850 JAG39754.1 GBHO01003851 JAG39753.1 GBRD01003659 JAG62162.1 GBHO01003847 JAG39757.1 GBHO01003846 JAG39758.1 GBHO01003845 JAG39759.1 GBHO01003848 JAG39756.1 GBHO01009597 JAG34007.1 GBHO01003853 JAG39751.1 PNF20189.1 PNF20190.1 QOIP01000002 RLU25330.1 GBGD01000006 JAC88883.1 GFDF01000226 JAV13858.1 GFTR01009002 JAW07424.1 GBBI01003645 JAC15067.1 CH933806 KRG01228.1 EDW14826.1 CH479185 EDW38537.1 KRG01220.1 KRG01221.1 KRG01225.1 KRG01222.1 KRG01226.1 KRG01224.1 KRG01229.1 KRG01223.1 KRG01219.1 KRG01227.1 AE014297 AGB96113.1 CH916378 EDV98610.1 CH940650 EDW68106.1 KRF83673.1 KRF83657.1 KRF83660.1 KRF83666.1 KRF83661.1 KRF83663.1 KRF83668.1 KRF83656.1 AGB96110.1 KRF83671.1 KRF83665.1 KRF83659.1 KRF83667.1 AF277386 AF297644 AF300456 AF302236 AAF55673.2 AGB96112.1 CM000070 KRT01348.1 KRF83669.1 KRF83664.1 KRT01360.1 AF300294 X99757 AAF55676.3 AAK15257.1 KRF83670.1 KRF83672.1 KRT01345.1 CP012526 ALC46996.1 KRT01342.1 KRF83662.1 KRT01343.1 EAL29187.4
Proteomes
Pfam
Interpro
IPR018159
Spectrin/alpha-actinin
+ More
IPR002017 Spectrin_repeat
IPR011992 EF-hand-dom_pair
IPR001202 WW_dom
IPR036020 WW_dom_sf
IPR000433 Znf_ZZ
IPR015153 EF-hand_dom_typ1
IPR015154 EF-hand_dom_typ2
IPR001750 ND/Mrp_mem
IPR035436 Dystrophin/utrophin
IPR036872 CH_dom_sf
IPR001589 Actinin_actin-bd_CS
IPR001715 CH-domain
IPR002017 Spectrin_repeat
IPR011992 EF-hand-dom_pair
IPR001202 WW_dom
IPR036020 WW_dom_sf
IPR000433 Znf_ZZ
IPR015153 EF-hand_dom_typ1
IPR015154 EF-hand_dom_typ2
IPR001750 ND/Mrp_mem
IPR035436 Dystrophin/utrophin
IPR036872 CH_dom_sf
IPR001589 Actinin_actin-bd_CS
IPR001715 CH-domain
Gene 3D
CDD
ProteinModelPortal
A0A212EQW8
A0A194PWC9
A0A1E1WJ00
A0A194RDD7
A0A1B6EW19
A0A1B6KWY7
+ More
A0A1B6LPQ0 A0A088AJ44 A0A1B6I1K0 A0A1B6IKG4 A0A310STA5 A0A232FMZ1 A0A1B6LEA1 A0A1B6KJ06 A0A158NE51 V9ICT2 A0A2A3EC27 E0VJZ9 A0A1B6HMA6 A0A0C9RVM7 A0A0L7R7U4 A0A0N0BDW7 A0A1B0G180 A0A336MUW4 A0A1A9X1L7 A0A1A9XP77 A0A336MMA3 A0A0K8TBX1 A0A0K8T9U8 A0A0K8T9F1 A0A0K8T9F7 A0A0K8TAJ4 A0A0A9YSD3 A0A0A9ZD72 A0A0A9YQV7 A0A0A9ZD78 A0A2J7PV18 A0A0A9Z5J2 A0A0A9Z8S5 A0A0K8T9D3 A0A0A9Z3C7 A0A0A9Z8T0 A0A0A9Z5J6 A0A0A9Z770 A0A0A9YLR0 A0A0A9Z766 A0A2J7PV24 A0A2J7PV58 A0A3L8DXZ2 A0A069DY06 A0A1L8E575 A0A224X947 A0A023F0P7 A0A0Q9WZ85 B4KCK7 B4GLW3 A0A0Q9X7F0 A0A0Q9X128 A0A0Q9WZ57 A0A0Q9X5Z0 A0A0Q9XB81 A0A0Q9WZP4 A0A0Q9X9Z4 A0A0Q9XBK8 A0A0Q9WZ87 A0A0B4KGB9 B4JYZ5 B4LZ03 A0A0Q9WGJ9 A0A0Q9WHH3 A0A0Q9WGH2 A0A0Q9WRQ6 A0A0Q9WSC2 A0A0Q9WGP2 A0A0B4KHR4 A0A0Q9WNZ9 A0A1W4W9F8 A0A0Q9WQI2 A0A0Q9WSU5 A0A0Q9WH19 Q9VDW6-2 A0A0B4KGH0 A0A0R3NKF7 A0A0Q9WU35 A0A0Q9WU41 A0A0R3NR30 Q9VDW3 A0A0Q9WIB5 A0A0Q9WGQ7 A0A0R3NK00 A0A0M4EJD4 A0A0R3NKA5 A0A0Q9WSF8 A0A0R3NPN5 Q9VDW6 A0A1W4W848 Q293M7
A0A1B6LPQ0 A0A088AJ44 A0A1B6I1K0 A0A1B6IKG4 A0A310STA5 A0A232FMZ1 A0A1B6LEA1 A0A1B6KJ06 A0A158NE51 V9ICT2 A0A2A3EC27 E0VJZ9 A0A1B6HMA6 A0A0C9RVM7 A0A0L7R7U4 A0A0N0BDW7 A0A1B0G180 A0A336MUW4 A0A1A9X1L7 A0A1A9XP77 A0A336MMA3 A0A0K8TBX1 A0A0K8T9U8 A0A0K8T9F1 A0A0K8T9F7 A0A0K8TAJ4 A0A0A9YSD3 A0A0A9ZD72 A0A0A9YQV7 A0A0A9ZD78 A0A2J7PV18 A0A0A9Z5J2 A0A0A9Z8S5 A0A0K8T9D3 A0A0A9Z3C7 A0A0A9Z8T0 A0A0A9Z5J6 A0A0A9Z770 A0A0A9YLR0 A0A0A9Z766 A0A2J7PV24 A0A2J7PV58 A0A3L8DXZ2 A0A069DY06 A0A1L8E575 A0A224X947 A0A023F0P7 A0A0Q9WZ85 B4KCK7 B4GLW3 A0A0Q9X7F0 A0A0Q9X128 A0A0Q9WZ57 A0A0Q9X5Z0 A0A0Q9XB81 A0A0Q9WZP4 A0A0Q9X9Z4 A0A0Q9XBK8 A0A0Q9WZ87 A0A0B4KGB9 B4JYZ5 B4LZ03 A0A0Q9WGJ9 A0A0Q9WHH3 A0A0Q9WGH2 A0A0Q9WRQ6 A0A0Q9WSC2 A0A0Q9WGP2 A0A0B4KHR4 A0A0Q9WNZ9 A0A1W4W9F8 A0A0Q9WQI2 A0A0Q9WSU5 A0A0Q9WH19 Q9VDW6-2 A0A0B4KGH0 A0A0R3NKF7 A0A0Q9WU35 A0A0Q9WU41 A0A0R3NR30 Q9VDW3 A0A0Q9WIB5 A0A0Q9WGQ7 A0A0R3NK00 A0A0M4EJD4 A0A0R3NKA5 A0A0Q9WSF8 A0A0R3NPN5 Q9VDW6 A0A1W4W848 Q293M7
Ontologies
GO
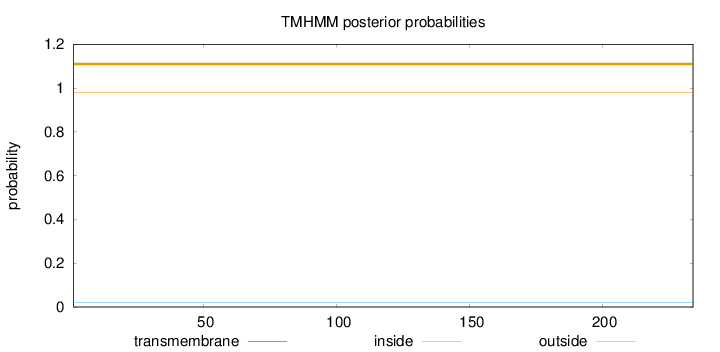
Topology
Subcellular location
Cell membrane
Sarcolemma
Cytoplasm
Cytoskeleton
Sarcolemma
Cytoplasm
Cytoskeleton
Length:
234
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8e-05
Exp number, first 60 AAs:
8e-05
Total prob of N-in:
0.01980
outside
1 - 234
Population Genetic Test Statistics
Pi
206.433919
Theta
164.816211
Tajima's D
0.045668
CLR
0.511106
CSRT
0.379531023448828
Interpretation
Possibly Positive selection
