Gene
KWMTBOMO04201
Pre Gene Modal
BGIBMGA008648
Annotation
PREDICTED:_dystrophin?_isoform_B-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.061
Sequence
CDS
ATGGAGGAACCACAAGGTTGGCTCTCGCACTGCGACAAGCGCGTACGAGCCTTCACCAACTCTCTTGACGAGCTCGCCAGCAGAGTCCAAGCTGCTGAAGCGGCCAGGTCTTCATGGCGAGGACCAGGCGACGCAAGAGACGCTCGGGCACAACTGGATGCTGTCTCCAGAGCCAGAGCTCAACTGCCACCACTCAGGAGATTGGCTGATGAGCTACACGCTCAAGCACAGACACTAGCGAGAGACAAAATCCAGCTACCAGATCAATTACTGGCGAAACTTGATGACTTAAATACAAGAGTCGGTGCTCTCTGCGCTGGTGGGGAGGAACGGGCCCGTCAGCTGGCAGGCGTGGCCCGCGACGGTGGGGCCGGCGCTGCCCAGGGATTCCTCGCTGGTTCCGTGGACCCACCGTGGGAGCGAGCCGTCACGCCAGCCAACGTACCTTATTATATTAACCATGAGTTAGAGACAACGCACTGGGACCACCCCAAGATGATTGAACTAATGAACTCCTTAGCCGACCTCAACGAGGTTCGGTTCTCGGCCTACAGGACCGCTCTCAAGCTACGGACGGTGCAGAAAGCTCTGTGCATGCACATGCTGCAGCTCCCGGCCGCGCTCGAGGCTTTCGACTCGCACGGTCTACGCGCGCAAAACGACAGACTCATCGACATTCCCGACATGATCACCGTCCTCACCACTCTGTACGAAGTGATTGCGGCGGAGAACCCAAGCGCCGTGAACGTGCCGCTGTGTCTGGACCTCTCCATGAACTGGCTGCTGAACGTGTACGACAGCCAGCGCACCGGACAGATCAGGGTGTTGAGCTTCAAAGTCGGCTTGGTGTTGCTCTGTAAAGGACATCTGGAGGAAAAGTATAGGTATTTATTCCGGTTAATCGCCGACCCATCATGCAGGGCAGACCAAAGGAAGCTGGGTCTCTTGCTGCACGATTGTATTCAGGTGCCGAGACAGTTGGGCGAGGTTGCAGCTTTCGGCGGCTCCAATATCGAACCCTCAGTCCGGAGTTGCTTCGAGCAGGCCTCCACTGCTGCCAAGGAGGGCAGCAAAGGAGGAACTCTTGACAGGAAAACTGAAAAAGAAAAGGATGAAACGAAAGAGAAGCCAGTCGAGCGAGACGCGGACGGCCAGGTCCAGTACATCGAAGCGTTCCAGTTCCTGCAGTGGCTGCAGCGCGAGCCGCAGTCCATGGTGTGGCTGCCCGTGCTGCACCGCCTGGCCGCCGCCGAGGGGGCCCGCCACCAAGCCAAGTGCAACATCTGCAAGGACTATCCTATCGTTGGGTTCAGGTACCGCTGCCTCAAGTGCTTCAACTTCGACATGTGTCAAAAGTGCTTCTTCAACGGCCGGAAGGCCAAGAACCACAAATTGACGCATCCGATGCAGGAGTACTGCACTGCGACGACGTCGGGAGAGGACGTGCGCGACTTCACGAGGGCGCTCCGCAACAAGTTCAAGTCGGCGCGCTACTTCAAGAAGCACCCCAGGGTCGGGTACTTGCCTGTACAAACCGTTTTAGAAGGTGATGCTCTGGAGTCCCCCGCTCCCTCTCCGCAGCACGGCGCGTCCCTCGGCCCCGACATGCACTCGCGCCTCGAGCTGTACGCCACCCGCCTCGCGCAGGTCGAGCTCAGCGCCGCCGACACTCCCGACAGCGATGAAGAACATCAGCTCATAGCTCAGTACTGCGCCTCACTGAACGGCGGAGGGGAGTCCGAAGAAGCTCCTCGCTCGCCTGTGCAAGTGGTCTCCATTATCCACCGCGAGCAGAGACACGAACTCGAGGCCATGATTAGGGAACTGGAAGAAGAAAACGCAAGTCTCCAAGCGGAGTACGAGCGTCTGAAGGCTAAGCAGACTCCAGGCTCCACGCCCGAGGAGCAGCAGCACGGCGTCAGCGACAATAGCCGCAGCGGACCGGTAGACCAGGACATGATGGCCGAGGCTCGTCTTCTCCGTCAGCACAAGGGCCGGCTGGAGGCTCGCATGCACATTCTGGAGGAACACAACAGACAACTCGAAGCTCAGCTGCAGCGTCTACGCACCTTACTCGACGAGCCGGCCCAGGGCTCGCCCCCAGCGCGCACCGGAACATTACAGACACGTTCGGTGACCGCTTCGCAGCTTGCAACTGATTCACCAGCCAAAATGCACAATGGACTCTACCACGAGTCACAAACAAATGGCGGCGCTCGTTGGGCTGGCGCCGAGCGGCCCCCCCCGCCCCCGCACACCGTGCACTCCCTCATGCACTGCGCAGATGACTTGGGCCGTGCCGTAGAAGAGCTCGTCTCGGTGATGTCCGAGGACTCCGGCACGCCGAGCAACGCGCCACCGCAGTACACGAACGGGAAACCGGCAAACAACGAATGA
Protein
MEEPQGWLSHCDKRVRAFTNSLDELASRVQAAEAARSSWRGPGDARDARAQLDAVSRARAQLPPLRRLADELHAQAQTLARDKIQLPDQLLAKLDDLNTRVGALCAGGEERARQLAGVARDGGAGAAQGFLAGSVDPPWERAVTPANVPYYINHELETTHWDHPKMIELMNSLADLNEVRFSAYRTALKLRTVQKALCMHMLQLPAALEAFDSHGLRAQNDRLIDIPDMITVLTTLYEVIAAENPSAVNVPLCLDLSMNWLLNVYDSQRTGQIRVLSFKVGLVLLCKGHLEEKYRYLFRLIADPSCRADQRKLGLLLHDCIQVPRQLGEVAAFGGSNIEPSVRSCFEQASTAAKEGSKGGTLDRKTEKEKDETKEKPVERDADGQVQYIEAFQFLQWLQREPQSMVWLPVLHRLAAAEGARHQAKCNICKDYPIVGFRYRCLKCFNFDMCQKCFFNGRKAKNHKLTHPMQEYCTATTSGEDVRDFTRALRNKFKSARYFKKHPRVGYLPVQTVLEGDALESPAPSPQHGASLGPDMHSRLELYATRLAQVELSAADTPDSDEEHQLIAQYCASLNGGGESEEAPRSPVQVVSIIHREQRHELEAMIRELEEENASLQAEYERLKAKQTPGSTPEEQQHGVSDNSRSGPVDQDMMAEARLLRQHKGRLEARMHILEEHNRQLEAQLQRLRTLLDEPAQGSPPARTGTLQTRSVTASQLATDSPAKMHNGLYHESQTNGGARWAGAERPPPPPHTVHSLMHCADDLGRAVEELVSVMSEDSGTPSNAPPQYTNGKPANNE
Summary
Uniprot
A0A2H1VIY5
A0A2W1BY66
A0A1E1WUH3
A0A212ER20
A0A1E1W3G0
A0A194PX88
+ More
A0A195D7N4 A0A195EYG8 A0A151X450 A0A158NE51 E9J516 A0A2J7PV58 A0A154PH74 A0A0L7R870 A0A195BR01 F4W825 A0A310SEN3 A0A195CLI0 A0A088AJ46 E2B8D5 A0A026WH37 V9ICT2 A0A2A3EC27 A0A2J7PV18 A0A0C9RVM7 A0A2J7PV24 A0A0C9RBW3 A0A0C9QUT0 A0A2A4JAJ2 A0A2A3EDD6 A0A0C9RFK4 A0A0N0BDW7 A0A232FMZ6 A0A1B6HWQ9 A0A1B6DAH2 A0A1Q3FU53 A0A1B6HGJ5 A0A3L8DXZ2 A0A1B6FA46 B0X0E8 A0A0P6IXE5 A0A067RB92 A0A023EUZ7 A0A0A9YLR0 A0A0A9Z766 E0VJZ9 Q175I9 A0A0A9ZD78 A0A0A9ZD72 A0A069DY06 A0A224X947 A0A023F0P7 A0A0K8T9F1 W4VR82 A0A0K8T9F7 A0A336MTP1 A0A0K8T9D3 A0A1L8E5U7 W8B6P5 W8AXC5 W8AC76 W8AIH3 A0A1I8MJ19 A0A0K8W373 T1PM18 A0A0A1WU22 A0A034V0N6 A0A0A1WQW4 A0A0A9Z770 A0A1I8PLW7 A0A0A1X3X5 A0A0A1XNJ6 A0A3B0JNE1 A0A034V2T5 A0A0A9Z5J2 A0A1J1HVV4 A0A0A9Z5J6 A0A0A9Z3C7 T1P855 A0A0K8VES8 A0A0A1WM29 A0A0A9Z8S5 A0A0A9Z8T0 W5JVQ4 A0A0L0CIP1 A0A0A9YSD3 A0A0R3NPN5 A0A0P8Y0C2 A0A1W4VX52 A0A0P8Y0C3 A0A0A9YQV7 A0A182JPI6 A0A0P9AS37 A0A0P8YN77 A0A0B4KGH0 A0A1W4VX22 A0A1W4W848 A0A0R3NKE7 A0A0R3NQL5 A0A0R3NR30
A0A195D7N4 A0A195EYG8 A0A151X450 A0A158NE51 E9J516 A0A2J7PV58 A0A154PH74 A0A0L7R870 A0A195BR01 F4W825 A0A310SEN3 A0A195CLI0 A0A088AJ46 E2B8D5 A0A026WH37 V9ICT2 A0A2A3EC27 A0A2J7PV18 A0A0C9RVM7 A0A2J7PV24 A0A0C9RBW3 A0A0C9QUT0 A0A2A4JAJ2 A0A2A3EDD6 A0A0C9RFK4 A0A0N0BDW7 A0A232FMZ6 A0A1B6HWQ9 A0A1B6DAH2 A0A1Q3FU53 A0A1B6HGJ5 A0A3L8DXZ2 A0A1B6FA46 B0X0E8 A0A0P6IXE5 A0A067RB92 A0A023EUZ7 A0A0A9YLR0 A0A0A9Z766 E0VJZ9 Q175I9 A0A0A9ZD78 A0A0A9ZD72 A0A069DY06 A0A224X947 A0A023F0P7 A0A0K8T9F1 W4VR82 A0A0K8T9F7 A0A336MTP1 A0A0K8T9D3 A0A1L8E5U7 W8B6P5 W8AXC5 W8AC76 W8AIH3 A0A1I8MJ19 A0A0K8W373 T1PM18 A0A0A1WU22 A0A034V0N6 A0A0A1WQW4 A0A0A9Z770 A0A1I8PLW7 A0A0A1X3X5 A0A0A1XNJ6 A0A3B0JNE1 A0A034V2T5 A0A0A9Z5J2 A0A1J1HVV4 A0A0A9Z5J6 A0A0A9Z3C7 T1P855 A0A0K8VES8 A0A0A1WM29 A0A0A9Z8S5 A0A0A9Z8T0 W5JVQ4 A0A0L0CIP1 A0A0A9YSD3 A0A0R3NPN5 A0A0P8Y0C2 A0A1W4VX52 A0A0P8Y0C3 A0A0A9YQV7 A0A182JPI6 A0A0P9AS37 A0A0P8YN77 A0A0B4KGH0 A0A1W4VX22 A0A1W4W848 A0A0R3NKE7 A0A0R3NQL5 A0A0R3NR30
Pubmed
28756777
22118469
26354079
21347285
21282665
21719571
+ More
20798317 24508170 28648823 30249741 26999592 24845553 24945155 25401762 20566863 17510324 26334808 25474469 24495485 25315136 25830018 25348373 20920257 23761445 26108605 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
20798317 24508170 28648823 30249741 26999592 24845553 24945155 25401762 20566863 17510324 26334808 25474469 24495485 25315136 25830018 25348373 20920257 23761445 26108605 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
ODYU01002550
SOQ40204.1
KZ149948
PZC76713.1
GDQN01000371
JAT90683.1
+ More
AGBW02013189 OWR43894.1 GDQN01009536 JAT81518.1 KQ459589 KPI97633.1 KQ981153 KYN08886.1 KQ981906 KYN33263.1 KQ982548 KYQ55193.1 ADTU01012837 ADTU01012838 ADTU01012839 ADTU01012840 ADTU01012841 ADTU01012842 ADTU01012843 ADTU01012844 ADTU01012845 ADTU01012846 GL768123 EFZ12083.1 NEVH01020989 PNF20190.1 KQ434899 KZC10824.1 KQ414637 KOC66951.1 KQ976424 KYM88389.1 GL887888 EGI69695.1 KQ760629 OAD59720.1 KQ977600 KYN01563.1 GL446312 EFN88034.1 KK107207 EZA55377.1 JR038061 AEY58099.1 KZ288290 PBC29240.1 PNF20188.1 GBYB01011751 JAG81518.1 PNF20189.1 GBYB01004391 JAG74158.1 GBYB01004392 JAG74159.1 NWSH01002258 PCG68786.1 PBC29239.1 GBYB01007170 JAG76937.1 KQ435851 KOX70714.1 NNAY01000019 OXU31890.1 GECU01028599 JAS79107.1 GEDC01014605 JAS22693.1 GFDL01003977 JAV31068.1 GECU01033967 JAS73739.1 QOIP01000002 RLU25330.1 GECZ01022734 JAS47035.1 DS232235 EDS38109.1 GDUN01000260 JAN95659.1 KK852759 KDR17040.1 GAPW01000323 JAC13275.1 GBHO01009597 JAG34007.1 GBHO01003853 JAG39751.1 DS235235 EEB13705.1 CH477400 EAT41731.1 GBHO01003849 JAG39755.1 GBHO01003854 JAG39750.1 GBGD01000006 JAC88883.1 GFTR01009002 JAW07424.1 GBBI01003645 JAC15067.1 GBRD01003657 JAG62164.1 GANO01003961 JAB55910.1 GBRD01003638 JAG62183.1 UFQT01002627 SSX33802.1 GBRD01003659 JAG62162.1 GFDF01000169 JAV13915.1 GAMC01021106 JAB85449.1 GAMC01021104 JAB85451.1 GAMC01021105 JAB85450.1 GAMC01021108 JAB85447.1 GDHF01006979 JAI45335.1 KA648958 AFP63587.1 GBXI01011718 JAD02574.1 GAKP01022136 JAC36816.1 GBXI01013241 JAD01051.1 GBHO01003848 JAG39756.1 GBXI01008510 JAD05782.1 GBXI01001765 JAD12527.1 OUUW01000008 SPP83747.1 GAKP01022138 JAC36814.1 GBHO01003850 JAG39754.1 CVRI01000024 CRK92192.1 GBHO01003845 JAG39759.1 GBHO01003847 JAG39757.1 KA644762 AFP59391.1 GDHF01014946 JAI37368.1 GBXI01014839 JAC99452.1 GBHO01003851 JAG39753.1 GBHO01003846 JAG39758.1 ADMH02000278 ETN67140.1 JRES01000342 KNC32077.1 GBHO01009596 JAG34008.1 CM000070 KRT01343.1 CH902617 KPU80241.1 KPU80239.1 GBHO01009598 JAG34006.1 KPU80240.1 KPU80243.1 AE014297 AGB96112.1 KRT01355.1 KRT01349.1 KRT01360.1
AGBW02013189 OWR43894.1 GDQN01009536 JAT81518.1 KQ459589 KPI97633.1 KQ981153 KYN08886.1 KQ981906 KYN33263.1 KQ982548 KYQ55193.1 ADTU01012837 ADTU01012838 ADTU01012839 ADTU01012840 ADTU01012841 ADTU01012842 ADTU01012843 ADTU01012844 ADTU01012845 ADTU01012846 GL768123 EFZ12083.1 NEVH01020989 PNF20190.1 KQ434899 KZC10824.1 KQ414637 KOC66951.1 KQ976424 KYM88389.1 GL887888 EGI69695.1 KQ760629 OAD59720.1 KQ977600 KYN01563.1 GL446312 EFN88034.1 KK107207 EZA55377.1 JR038061 AEY58099.1 KZ288290 PBC29240.1 PNF20188.1 GBYB01011751 JAG81518.1 PNF20189.1 GBYB01004391 JAG74158.1 GBYB01004392 JAG74159.1 NWSH01002258 PCG68786.1 PBC29239.1 GBYB01007170 JAG76937.1 KQ435851 KOX70714.1 NNAY01000019 OXU31890.1 GECU01028599 JAS79107.1 GEDC01014605 JAS22693.1 GFDL01003977 JAV31068.1 GECU01033967 JAS73739.1 QOIP01000002 RLU25330.1 GECZ01022734 JAS47035.1 DS232235 EDS38109.1 GDUN01000260 JAN95659.1 KK852759 KDR17040.1 GAPW01000323 JAC13275.1 GBHO01009597 JAG34007.1 GBHO01003853 JAG39751.1 DS235235 EEB13705.1 CH477400 EAT41731.1 GBHO01003849 JAG39755.1 GBHO01003854 JAG39750.1 GBGD01000006 JAC88883.1 GFTR01009002 JAW07424.1 GBBI01003645 JAC15067.1 GBRD01003657 JAG62164.1 GANO01003961 JAB55910.1 GBRD01003638 JAG62183.1 UFQT01002627 SSX33802.1 GBRD01003659 JAG62162.1 GFDF01000169 JAV13915.1 GAMC01021106 JAB85449.1 GAMC01021104 JAB85451.1 GAMC01021105 JAB85450.1 GAMC01021108 JAB85447.1 GDHF01006979 JAI45335.1 KA648958 AFP63587.1 GBXI01011718 JAD02574.1 GAKP01022136 JAC36816.1 GBXI01013241 JAD01051.1 GBHO01003848 JAG39756.1 GBXI01008510 JAD05782.1 GBXI01001765 JAD12527.1 OUUW01000008 SPP83747.1 GAKP01022138 JAC36814.1 GBHO01003850 JAG39754.1 CVRI01000024 CRK92192.1 GBHO01003845 JAG39759.1 GBHO01003847 JAG39757.1 KA644762 AFP59391.1 GDHF01014946 JAI37368.1 GBXI01014839 JAC99452.1 GBHO01003851 JAG39753.1 GBHO01003846 JAG39758.1 ADMH02000278 ETN67140.1 JRES01000342 KNC32077.1 GBHO01009596 JAG34008.1 CM000070 KRT01343.1 CH902617 KPU80241.1 KPU80239.1 GBHO01009598 JAG34006.1 KPU80240.1 KPU80243.1 AE014297 AGB96112.1 KRT01355.1 KRT01349.1 KRT01360.1
Proteomes
UP000007151
UP000053268
UP000078492
UP000078541
UP000075809
UP000005205
+ More
UP000235965 UP000076502 UP000053825 UP000078540 UP000007755 UP000078542 UP000005203 UP000008237 UP000053097 UP000242457 UP000218220 UP000053105 UP000215335 UP000279307 UP000002320 UP000027135 UP000009046 UP000008820 UP000095301 UP000095300 UP000268350 UP000183832 UP000000673 UP000037069 UP000001819 UP000007801 UP000192221 UP000075881 UP000000803
UP000235965 UP000076502 UP000053825 UP000078540 UP000007755 UP000078542 UP000005203 UP000008237 UP000053097 UP000242457 UP000218220 UP000053105 UP000215335 UP000279307 UP000002320 UP000027135 UP000009046 UP000008820 UP000095301 UP000095300 UP000268350 UP000183832 UP000000673 UP000037069 UP000001819 UP000007801 UP000192221 UP000075881 UP000000803
Pfam
Interpro
IPR000433
Znf_ZZ
+ More
IPR036020 WW_dom_sf
IPR001202 WW_dom
IPR011992 EF-hand-dom_pair
IPR015154 EF-hand_dom_typ2
IPR018159 Spectrin/alpha-actinin
IPR015153 EF-hand_dom_typ1
IPR002017 Spectrin_repeat
IPR001750 ND/Mrp_mem
IPR035436 Dystrophin/utrophin
IPR001589 Actinin_actin-bd_CS
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR036020 WW_dom_sf
IPR001202 WW_dom
IPR011992 EF-hand-dom_pair
IPR015154 EF-hand_dom_typ2
IPR018159 Spectrin/alpha-actinin
IPR015153 EF-hand_dom_typ1
IPR002017 Spectrin_repeat
IPR001750 ND/Mrp_mem
IPR035436 Dystrophin/utrophin
IPR001589 Actinin_actin-bd_CS
IPR001715 CH-domain
IPR036872 CH_dom_sf
Gene 3D
CDD
ProteinModelPortal
A0A2H1VIY5
A0A2W1BY66
A0A1E1WUH3
A0A212ER20
A0A1E1W3G0
A0A194PX88
+ More
A0A195D7N4 A0A195EYG8 A0A151X450 A0A158NE51 E9J516 A0A2J7PV58 A0A154PH74 A0A0L7R870 A0A195BR01 F4W825 A0A310SEN3 A0A195CLI0 A0A088AJ46 E2B8D5 A0A026WH37 V9ICT2 A0A2A3EC27 A0A2J7PV18 A0A0C9RVM7 A0A2J7PV24 A0A0C9RBW3 A0A0C9QUT0 A0A2A4JAJ2 A0A2A3EDD6 A0A0C9RFK4 A0A0N0BDW7 A0A232FMZ6 A0A1B6HWQ9 A0A1B6DAH2 A0A1Q3FU53 A0A1B6HGJ5 A0A3L8DXZ2 A0A1B6FA46 B0X0E8 A0A0P6IXE5 A0A067RB92 A0A023EUZ7 A0A0A9YLR0 A0A0A9Z766 E0VJZ9 Q175I9 A0A0A9ZD78 A0A0A9ZD72 A0A069DY06 A0A224X947 A0A023F0P7 A0A0K8T9F1 W4VR82 A0A0K8T9F7 A0A336MTP1 A0A0K8T9D3 A0A1L8E5U7 W8B6P5 W8AXC5 W8AC76 W8AIH3 A0A1I8MJ19 A0A0K8W373 T1PM18 A0A0A1WU22 A0A034V0N6 A0A0A1WQW4 A0A0A9Z770 A0A1I8PLW7 A0A0A1X3X5 A0A0A1XNJ6 A0A3B0JNE1 A0A034V2T5 A0A0A9Z5J2 A0A1J1HVV4 A0A0A9Z5J6 A0A0A9Z3C7 T1P855 A0A0K8VES8 A0A0A1WM29 A0A0A9Z8S5 A0A0A9Z8T0 W5JVQ4 A0A0L0CIP1 A0A0A9YSD3 A0A0R3NPN5 A0A0P8Y0C2 A0A1W4VX52 A0A0P8Y0C3 A0A0A9YQV7 A0A182JPI6 A0A0P9AS37 A0A0P8YN77 A0A0B4KGH0 A0A1W4VX22 A0A1W4W848 A0A0R3NKE7 A0A0R3NQL5 A0A0R3NR30
A0A195D7N4 A0A195EYG8 A0A151X450 A0A158NE51 E9J516 A0A2J7PV58 A0A154PH74 A0A0L7R870 A0A195BR01 F4W825 A0A310SEN3 A0A195CLI0 A0A088AJ46 E2B8D5 A0A026WH37 V9ICT2 A0A2A3EC27 A0A2J7PV18 A0A0C9RVM7 A0A2J7PV24 A0A0C9RBW3 A0A0C9QUT0 A0A2A4JAJ2 A0A2A3EDD6 A0A0C9RFK4 A0A0N0BDW7 A0A232FMZ6 A0A1B6HWQ9 A0A1B6DAH2 A0A1Q3FU53 A0A1B6HGJ5 A0A3L8DXZ2 A0A1B6FA46 B0X0E8 A0A0P6IXE5 A0A067RB92 A0A023EUZ7 A0A0A9YLR0 A0A0A9Z766 E0VJZ9 Q175I9 A0A0A9ZD78 A0A0A9ZD72 A0A069DY06 A0A224X947 A0A023F0P7 A0A0K8T9F1 W4VR82 A0A0K8T9F7 A0A336MTP1 A0A0K8T9D3 A0A1L8E5U7 W8B6P5 W8AXC5 W8AC76 W8AIH3 A0A1I8MJ19 A0A0K8W373 T1PM18 A0A0A1WU22 A0A034V0N6 A0A0A1WQW4 A0A0A9Z770 A0A1I8PLW7 A0A0A1X3X5 A0A0A1XNJ6 A0A3B0JNE1 A0A034V2T5 A0A0A9Z5J2 A0A1J1HVV4 A0A0A9Z5J6 A0A0A9Z3C7 T1P855 A0A0K8VES8 A0A0A1WM29 A0A0A9Z8S5 A0A0A9Z8T0 W5JVQ4 A0A0L0CIP1 A0A0A9YSD3 A0A0R3NPN5 A0A0P8Y0C2 A0A1W4VX52 A0A0P8Y0C3 A0A0A9YQV7 A0A182JPI6 A0A0P9AS37 A0A0P8YN77 A0A0B4KGH0 A0A1W4VX22 A0A1W4W848 A0A0R3NKE7 A0A0R3NQL5 A0A0R3NR30
PDB
1EG4
E-value=1.64229e-90,
Score=852
Ontologies
GO
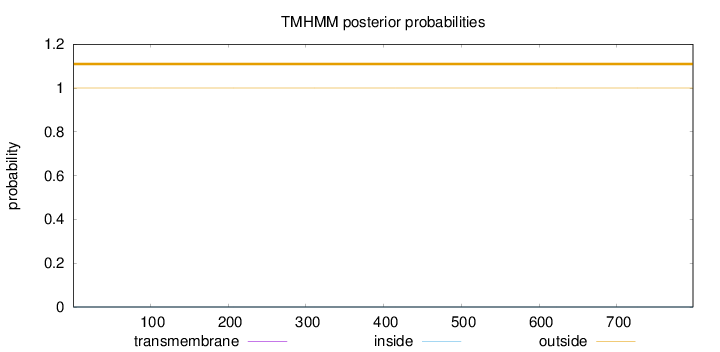
Topology
Length:
798
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 798
Population Genetic Test Statistics
Pi
291.048819
Theta
182.960114
Tajima's D
1.620345
CLR
0.224071
CSRT
0.812959352032398
Interpretation
Uncertain
