Pre Gene Modal
BGIBMGA008650
Annotation
cyclin-dependent_kinase_regulatory_subunit_[Bombyx_mori]
Full name
Cyclin-dependent kinases regulatory subunit
Location in the cell
Mitochondrial Reliability : 1.017 Nuclear Reliability : 1.368
Sequence
CDS
ATGCCCGTGGACCAGATTCAATATTCGGAAAGATATACAGATGACGTCTACGAATACAGGCATGTCATCTTACCACCGGATATAGCCCGCATGGTGCCAAAATCCCATCTTATGACAGAGACAGAGTGGCGAAACCTTGGTGTTCAGCAGAGTCCCGGCTGGCTCCACTTTATGGTGCATAATCCTGAACCCCATGTTCTATTATTTAGAAGACCTAGAACTGATATACCGGCTCCCTTGAATGGCTTAGATACAAATACTAGTTCTGCTGTGAAGTTGATTACGAAATTGGCTTGGCTGCAAACGCGAAACTAA
Protein
MPVDQIQYSERYTDDVYEYRHVILPPDIARMVPKSHLMTETEWRNLGVQQSPGWLHFMVHNPEPHVLLFRRPRTDIPAPLNGLDTNTSSAVKLITKLAWLQTRN
Summary
Description
Binds to the catalytic subunit of the cyclin dependent kinases and is essential for their biological function.
Similarity
Belongs to the CKS family.
Uniprot
H9JGK3
Q2F6C0
A0A2A4J9M4
A0A0B4KK24
A0A212EQW5
S4PWJ6
+ More
A0A2H1VH89 A0A0L7LTU2 A0A194PY07 A0A194RCY5 A0A034WTE9 A0A0A1X3B8 A0A0K8UI31 A0A023EEI9 Q17AP8 A0A232EJX9 A0A1Q3FDX1 A0A0L0C923 A0A3L8D691 A0A1A9WPT4 A0A067R430 A0A2J7QT56 K7IXS1 A0A1I8PJF2 A0A023EE40 A0A182WV61 Q7QFX2 A0A182I8T6 A0A1I8N8F9 A0A182NL45 B0WH04 A0A2A3E959 A0A088A8A9 A0A1A9Z868 A0A1A9XM48 A0A1B0BQB5 A0A182P789 W8BDG0 D3TPR3 A0A1A9VDG8 U5ETE0 A0A182R5W9 A0A2P8XWX3 D1FQ33 A0A310SHA7 A0A0K8TNY1 F4WWH9 A0A195FCT3 A0A0N1ITP1 A0A195BNF4 A0A158NKN9 B4JI00 A0A182F9H1 A0A2M4C526 B4K7J7 B4HL28 B4PTU2 B3P4U3 A0A1W4W2T5 Q9VHN1 B3LXG6 B4NB03 B4LZG5 A0A182XXE3 A0A3B0KCW2 Q294K5 B4GMZ3 A0A2M4CLT7 A0A1L8DKQ3 A0A336LNA2 J3JTS3 D0QWL8 A0A1B0DLP6 A0A1W4XWT4 A0A1Q3EXP6 A0A2B4REK6 D2A136 A0A1B0CJ40 E0VTD9 A0A3M6UVM4 A0A094ZG01 A0A183NZP3 A0A183JXZ4 A0A183MF79 A0A075AF98 Q5BSH4 C1LJY4 A0A183RDG0 G4VHW3
A0A2H1VH89 A0A0L7LTU2 A0A194PY07 A0A194RCY5 A0A034WTE9 A0A0A1X3B8 A0A0K8UI31 A0A023EEI9 Q17AP8 A0A232EJX9 A0A1Q3FDX1 A0A0L0C923 A0A3L8D691 A0A1A9WPT4 A0A067R430 A0A2J7QT56 K7IXS1 A0A1I8PJF2 A0A023EE40 A0A182WV61 Q7QFX2 A0A182I8T6 A0A1I8N8F9 A0A182NL45 B0WH04 A0A2A3E959 A0A088A8A9 A0A1A9Z868 A0A1A9XM48 A0A1B0BQB5 A0A182P789 W8BDG0 D3TPR3 A0A1A9VDG8 U5ETE0 A0A182R5W9 A0A2P8XWX3 D1FQ33 A0A310SHA7 A0A0K8TNY1 F4WWH9 A0A195FCT3 A0A0N1ITP1 A0A195BNF4 A0A158NKN9 B4JI00 A0A182F9H1 A0A2M4C526 B4K7J7 B4HL28 B4PTU2 B3P4U3 A0A1W4W2T5 Q9VHN1 B3LXG6 B4NB03 B4LZG5 A0A182XXE3 A0A3B0KCW2 Q294K5 B4GMZ3 A0A2M4CLT7 A0A1L8DKQ3 A0A336LNA2 J3JTS3 D0QWL8 A0A1B0DLP6 A0A1W4XWT4 A0A1Q3EXP6 A0A2B4REK6 D2A136 A0A1B0CJ40 E0VTD9 A0A3M6UVM4 A0A094ZG01 A0A183NZP3 A0A183JXZ4 A0A183MF79 A0A075AF98 Q5BSH4 C1LJY4 A0A183RDG0 G4VHW3
Pubmed
19121390
28756777
22118469
23622113
26227816
26354079
+ More
25348373 25830018 24945155 26483478 17510324 28648823 26108605 30249741 24845553 20075255 12364791 14747013 17210077 25315136 24495485 20353571 29403074 26369729 21719571 21347285 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 15632085 22516182 23537049 19859648 18362917 19820115 20566863 30382153 22246508 16617374 22253936
25348373 25830018 24945155 26483478 17510324 28648823 26108605 30249741 24845553 20075255 12364791 14747013 17210077 25315136 24495485 20353571 29403074 26369729 21719571 21347285 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 15632085 22516182 23537049 19859648 18362917 19820115 20566863 30382153 22246508 16617374 22253936
EMBL
BABH01039110
DQ311152
ABD36097.1
NWSH01002258
PCG68785.1
KC242802
+ More
KZ149948 AGW00925.1 PZC76714.1 AGBW02013189 OWR43893.1 GAIX01006673 JAA85887.1 ODYU01002550 SOQ40203.1 JTDY01000101 KOB78862.1 KQ459589 KPI97634.1 KQ460367 KPJ15482.1 GAKP01001899 JAC57053.1 GBXI01009052 JAD05240.1 GDHF01031695 GDHF01025977 GDHF01010597 JAI20619.1 JAI26337.1 JAI41717.1 JXUM01007517 GAPW01006528 KQ560241 JAC07070.1 KXJ83642.1 CH477331 EAT43338.1 NNAY01003942 OXU18611.1 GFDL01009299 JAV25746.1 JRES01000752 KNC28755.1 QOIP01000013 RLU15358.1 KK852771 KDR16878.1 NEVH01011202 PNF31753.1 GAPW01006527 JAC07071.1 AAAB01008844 EAA06091.3 APCN01003117 DS231931 EDS27329.1 KZ288325 PBC28004.1 JXJN01018551 GAMC01009828 GAMC01009827 JAB96727.1 CCAG010012556 EZ423415 ADD19691.1 GANO01001963 JAB57908.1 PYGN01001224 PSN36510.1 EZ419933 ACZ28288.1 KQ766586 OAD53665.1 GDAI01001757 JAI15846.1 GL888406 EGI61521.1 KQ981693 KYN37854.1 KQ435768 KOX75224.1 KQ976439 KYM86731.1 ADTU01018942 CH916369 EDV92908.1 GGFJ01011281 MBW60422.1 CH933806 EDW15341.1 CH480815 EDW42992.1 CM000160 EDW96553.1 CH954181 EDV49887.1 AE014297 AY102666 AAF54272.1 AAM27495.1 CH902617 EDV42810.1 CH964232 EDW80967.1 CH940650 EDW68200.1 OUUW01000008 SPP84049.1 CM000070 EAL28963.1 CH479185 EDW38217.1 GGFL01002057 MBW66235.1 GFDF01007031 JAV07053.1 UFQS01000027 UFQT01000027 SSW97693.1 SSX18079.1 APGK01023056 BT126632 KB740462 AEE61595.1 ENN80606.1 FJ821033 ACN94717.1 AJVK01072439 GFDL01015067 JAV19978.1 LSMT01000669 PFX15239.1 KQ971338 EFA02600.1 AJWK01014019 DS235762 EEB16645.1 RCHS01000617 RMX57746.1 KL250514 KGB32722.1 UZAL01028320 VDP40292.1 UZAK01032399 VDP26912.1 UZAI01016813 VDP16471.1 KL596721 KER27504.1 AY915290 AAX30511.2 FN314628 FN319284 FN319285 CAX75012.1 HE601627 CCD79014.1
KZ149948 AGW00925.1 PZC76714.1 AGBW02013189 OWR43893.1 GAIX01006673 JAA85887.1 ODYU01002550 SOQ40203.1 JTDY01000101 KOB78862.1 KQ459589 KPI97634.1 KQ460367 KPJ15482.1 GAKP01001899 JAC57053.1 GBXI01009052 JAD05240.1 GDHF01031695 GDHF01025977 GDHF01010597 JAI20619.1 JAI26337.1 JAI41717.1 JXUM01007517 GAPW01006528 KQ560241 JAC07070.1 KXJ83642.1 CH477331 EAT43338.1 NNAY01003942 OXU18611.1 GFDL01009299 JAV25746.1 JRES01000752 KNC28755.1 QOIP01000013 RLU15358.1 KK852771 KDR16878.1 NEVH01011202 PNF31753.1 GAPW01006527 JAC07071.1 AAAB01008844 EAA06091.3 APCN01003117 DS231931 EDS27329.1 KZ288325 PBC28004.1 JXJN01018551 GAMC01009828 GAMC01009827 JAB96727.1 CCAG010012556 EZ423415 ADD19691.1 GANO01001963 JAB57908.1 PYGN01001224 PSN36510.1 EZ419933 ACZ28288.1 KQ766586 OAD53665.1 GDAI01001757 JAI15846.1 GL888406 EGI61521.1 KQ981693 KYN37854.1 KQ435768 KOX75224.1 KQ976439 KYM86731.1 ADTU01018942 CH916369 EDV92908.1 GGFJ01011281 MBW60422.1 CH933806 EDW15341.1 CH480815 EDW42992.1 CM000160 EDW96553.1 CH954181 EDV49887.1 AE014297 AY102666 AAF54272.1 AAM27495.1 CH902617 EDV42810.1 CH964232 EDW80967.1 CH940650 EDW68200.1 OUUW01000008 SPP84049.1 CM000070 EAL28963.1 CH479185 EDW38217.1 GGFL01002057 MBW66235.1 GFDF01007031 JAV07053.1 UFQS01000027 UFQT01000027 SSW97693.1 SSX18079.1 APGK01023056 BT126632 KB740462 AEE61595.1 ENN80606.1 FJ821033 ACN94717.1 AJVK01072439 GFDL01015067 JAV19978.1 LSMT01000669 PFX15239.1 KQ971338 EFA02600.1 AJWK01014019 DS235762 EEB16645.1 RCHS01000617 RMX57746.1 KL250514 KGB32722.1 UZAL01028320 VDP40292.1 UZAK01032399 VDP26912.1 UZAI01016813 VDP16471.1 KL596721 KER27504.1 AY915290 AAX30511.2 FN314628 FN319284 FN319285 CAX75012.1 HE601627 CCD79014.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053268
UP000053240
+ More
UP000069940 UP000249989 UP000008820 UP000215335 UP000037069 UP000279307 UP000091820 UP000027135 UP000235965 UP000002358 UP000095300 UP000076407 UP000007062 UP000075840 UP000095301 UP000075884 UP000002320 UP000242457 UP000005203 UP000092445 UP000092443 UP000092460 UP000075885 UP000092444 UP000078200 UP000075900 UP000245037 UP000007755 UP000078541 UP000053105 UP000078540 UP000005205 UP000001070 UP000069272 UP000009192 UP000001292 UP000002282 UP000008711 UP000192221 UP000000803 UP000007801 UP000007798 UP000008792 UP000076408 UP000268350 UP000001819 UP000008744 UP000019118 UP000092462 UP000192223 UP000225706 UP000007266 UP000092461 UP000009046 UP000275408 UP000050791 UP000050789 UP000050790 UP000277204 UP000050792 UP000008854
UP000069940 UP000249989 UP000008820 UP000215335 UP000037069 UP000279307 UP000091820 UP000027135 UP000235965 UP000002358 UP000095300 UP000076407 UP000007062 UP000075840 UP000095301 UP000075884 UP000002320 UP000242457 UP000005203 UP000092445 UP000092443 UP000092460 UP000075885 UP000092444 UP000078200 UP000075900 UP000245037 UP000007755 UP000078541 UP000053105 UP000078540 UP000005205 UP000001070 UP000069272 UP000009192 UP000001292 UP000002282 UP000008711 UP000192221 UP000000803 UP000007801 UP000007798 UP000008792 UP000076408 UP000268350 UP000001819 UP000008744 UP000019118 UP000092462 UP000192223 UP000225706 UP000007266 UP000092461 UP000009046 UP000275408 UP000050791 UP000050789 UP000050790 UP000277204 UP000050792 UP000008854
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JGK3
Q2F6C0
A0A2A4J9M4
A0A0B4KK24
A0A212EQW5
S4PWJ6
+ More
A0A2H1VH89 A0A0L7LTU2 A0A194PY07 A0A194RCY5 A0A034WTE9 A0A0A1X3B8 A0A0K8UI31 A0A023EEI9 Q17AP8 A0A232EJX9 A0A1Q3FDX1 A0A0L0C923 A0A3L8D691 A0A1A9WPT4 A0A067R430 A0A2J7QT56 K7IXS1 A0A1I8PJF2 A0A023EE40 A0A182WV61 Q7QFX2 A0A182I8T6 A0A1I8N8F9 A0A182NL45 B0WH04 A0A2A3E959 A0A088A8A9 A0A1A9Z868 A0A1A9XM48 A0A1B0BQB5 A0A182P789 W8BDG0 D3TPR3 A0A1A9VDG8 U5ETE0 A0A182R5W9 A0A2P8XWX3 D1FQ33 A0A310SHA7 A0A0K8TNY1 F4WWH9 A0A195FCT3 A0A0N1ITP1 A0A195BNF4 A0A158NKN9 B4JI00 A0A182F9H1 A0A2M4C526 B4K7J7 B4HL28 B4PTU2 B3P4U3 A0A1W4W2T5 Q9VHN1 B3LXG6 B4NB03 B4LZG5 A0A182XXE3 A0A3B0KCW2 Q294K5 B4GMZ3 A0A2M4CLT7 A0A1L8DKQ3 A0A336LNA2 J3JTS3 D0QWL8 A0A1B0DLP6 A0A1W4XWT4 A0A1Q3EXP6 A0A2B4REK6 D2A136 A0A1B0CJ40 E0VTD9 A0A3M6UVM4 A0A094ZG01 A0A183NZP3 A0A183JXZ4 A0A183MF79 A0A075AF98 Q5BSH4 C1LJY4 A0A183RDG0 G4VHW3
A0A2H1VH89 A0A0L7LTU2 A0A194PY07 A0A194RCY5 A0A034WTE9 A0A0A1X3B8 A0A0K8UI31 A0A023EEI9 Q17AP8 A0A232EJX9 A0A1Q3FDX1 A0A0L0C923 A0A3L8D691 A0A1A9WPT4 A0A067R430 A0A2J7QT56 K7IXS1 A0A1I8PJF2 A0A023EE40 A0A182WV61 Q7QFX2 A0A182I8T6 A0A1I8N8F9 A0A182NL45 B0WH04 A0A2A3E959 A0A088A8A9 A0A1A9Z868 A0A1A9XM48 A0A1B0BQB5 A0A182P789 W8BDG0 D3TPR3 A0A1A9VDG8 U5ETE0 A0A182R5W9 A0A2P8XWX3 D1FQ33 A0A310SHA7 A0A0K8TNY1 F4WWH9 A0A195FCT3 A0A0N1ITP1 A0A195BNF4 A0A158NKN9 B4JI00 A0A182F9H1 A0A2M4C526 B4K7J7 B4HL28 B4PTU2 B3P4U3 A0A1W4W2T5 Q9VHN1 B3LXG6 B4NB03 B4LZG5 A0A182XXE3 A0A3B0KCW2 Q294K5 B4GMZ3 A0A2M4CLT7 A0A1L8DKQ3 A0A336LNA2 J3JTS3 D0QWL8 A0A1B0DLP6 A0A1W4XWT4 A0A1Q3EXP6 A0A2B4REK6 D2A136 A0A1B0CJ40 E0VTD9 A0A3M6UVM4 A0A094ZG01 A0A183NZP3 A0A183JXZ4 A0A183MF79 A0A075AF98 Q5BSH4 C1LJY4 A0A183RDG0 G4VHW3
PDB
4YC6
E-value=1.1861e-24,
Score=273
Ontologies
GO
Topology
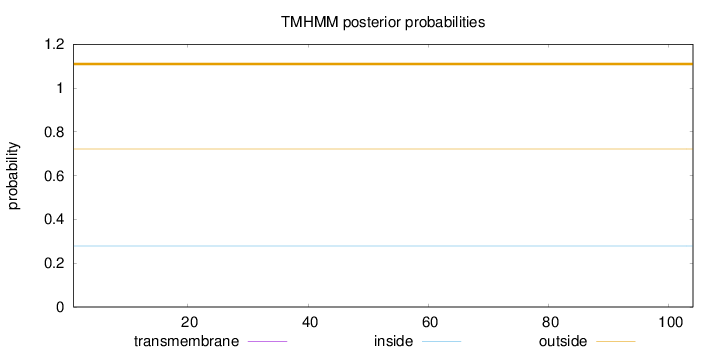
Length:
104
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00216
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.27846
outside
1 - 104
Population Genetic Test Statistics
Pi
250.197479
Theta
205.772839
Tajima's D
0.664636
CLR
141.160753
CSRT
0.56277186140693
Interpretation
Uncertain
