Pre Gene Modal
BGIBMGA008649
Annotation
PREDICTED:_uncharacterized_protein_LOC106135318_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.847
Sequence
CDS
ATGAGTGAGATCACGTTGGACTTCCTCCAGGCTCTGCTCAAAGATGAATATCCTGATGTCATTCTTCGTTCTTTCGAGGGAGCTCCCGGCTCGAAAAGAGGCGACAACTACACTTCGATGGTCTACAGGATCGCTTTGAAAGGAATCCAAAATCGGGTCGAACCGGACGGATCCTGCGAGACCGAAACACCATGGGAGGGGTCCATAATCTACAAAAGACTACCCGAGAGCACCACGAGGAGGGAAGCGTTCAAAAGCGACGAATTATTTTGCAACGAAATCGCGTTTTACAACAAGATCTGGCCGGCGCTATTCGAGTTTCAGAATCGATGGGGGATCGAGGAGCCATTCAAATCCGTACCTAAATGCTATTTAGCTAGGGATAACTGCGTTTTGCTGAAGGATCTGAAACAGTTCGGCTTCGTGATGCCCGATCGGAAAGTTGGATTGAGCGTGGACCAAACCTACTTCGTTCTGAGACATTTAGCGCACTTCCACGCTTTGTCATTGGCCATGAAGGGCCACGATCCGGAGGCTTTTCACGAGCTTTGGAATTCTAGGGACGGAATTTCTGAAGTGTTCTTCGTCTCCGAAAACGACGATTACTACCGAAACTACTACCACGAGGCCATCCAGAATGCCATAGCCATGGTGGACCAAGAGCTCCAGGGTACAGACTCTAAGGACCAATATCTGGACAGGTTCAAGGAATTTTGCAGCGAGGAGACTTTCTTTAAAACCATGGTGGAACTGGTCGAGCCTTGCGAACCTCTGGGCGTGATATGCCACGGAGACTGTTGGACTAACAACTTTCTCTTTCGATATGAAAATGAGAATATTGCGGAGATGTGCATCGTCGACTTCCAACTGGTTCGCTACGCGTCTCCAGCGTTGGACCTGGTATACATAATGTACCTGTGTCTGGAGCGACAGCAGCGAGCACTCCACTTGACCCCCCTGCTGCGGTACTACGCCGACCAGCTGTTCTGCAGGGCAGCGGAGCTCAGTGGCGGGGGTGTGGTGTGCGGGGGCGTTACCAGTGACGTGCTTTTTCAGATGCTCCACAAGGACTTCAGACAGAAGAGTCGCTTCGGTCTCGGCATAGCGCTGGACATGTACCCCATCACGACGTGCGACAGTGACGAGGCGCCCAACCTCTACTCAGCAGAGGACTCCGGGCGCTCCCAGGGCTCCGAGCCGGCGCCGGGTGCGCGTCCCGTGCGGACCTCCAGCGCGGCTTGCCGGAGGAAGATGACCGATCTGGTCAAAGAATTAGTCGACCAGGGTCTTTTGTAA
Protein
MSEITLDFLQALLKDEYPDVILRSFEGAPGSKRGDNYTSMVYRIALKGIQNRVEPDGSCETETPWEGSIIYKRLPESTTRREAFKSDELFCNEIAFYNKIWPALFEFQNRWGIEEPFKSVPKCYLARDNCVLLKDLKQFGFVMPDRKVGLSVDQTYFVLRHLAHFHALSLAMKGHDPEAFHELWNSRDGISEVFFVSENDDYYRNYYHEAIQNAIAMVDQELQGTDSKDQYLDRFKEFCSEETFFKTMVELVEPCEPLGVICHGDCWTNNFLFRYENENIAEMCIVDFQLVRYASPALDLVYIMYLCLERQQRALHLTPLLRYYADQLFCRAAELSGGGVVCGGVTSDVLFQMLHKDFRQKSRFGLGIALDMYPITTCDSDEAPNLYSAEDSGRSQGSEPAPGARPVRTSSAACRRKMTDLVKELVDQGLL
Summary
Uniprot
H9JGK2
A0A2H1VII8
A0A2A4JB62
A0A2W1BSV6
A0A3S2PLX5
A0A194RCB3
+ More
A0A212EQW9 A0A0L7LU10 A0A2P8YX69 A0A1B6KR03 A0A1B6DNL3 A0A1B6EHT1 A0A1B6IPS5 A0A2J7QLR3 E0W0A3 K7IWW7 A0A2S2PG70 A0A232EZP2 A0A182PFG1 A0A067RT58 E2BNI3 A0A182JML1 F4WR50 A0A182QQC1 W5JJE7 A0A151WEW2 A0A182FAA2 A0A084W4Y8 A0A182TYE4 A0A182XVL5 A0A195FKI4 E2AP81 A0A0L7QKP1 Q7PQT3 A0A182XEA8 A0A182N6Q7 A0A182V632 A0A182HJ12 A0A195BAS2 A0A158NU72 A0A151IQE1 A0A182K7P2 A0A1A9WWV3 A0A182RYE1 E9IHJ4 A0A1W7R8C1 A0A0K8VE71 A0A1B0EXN1 A0A182MGH5 A0A026WAF1 A0A1I8NNZ5 A0A154PF94 A0A1I8N754 A0A182W2U9 B3M2V1 B4LZ77 B0X9F7 A0A0M4EQU5 Q293U6 B4GLM0 A0A3B0JMJ9 B4KCC3 B4NB43 B4JUH7 W8BQE5 B3NZ98 A0A088AFG5 A0A1B6CFM7 Q9VEB7 B4IB91 A0A1W4U8E9 A0A310S7F4 A0A2A3EC89 V9IED8 B4PL80 A0A336L3U0 X1XCH0 A0A0K8U9F6 A0A182LC64 B4QUI4 A0A1B0D7X8 A0A2J7QMA2 Q17DW7 A0A023ERZ5 A0A182GTK0 Q7QBD5 A0A182HJA1 A0A182XE21 A0A1I8JUR4 A0A3F2Z0C7 A0A182S5M1 A0A2A4JMI3 A0A1Q3FS08 W8BSE4 A0A182K9D3 A0A182N5A7 A0A182PE46
A0A212EQW9 A0A0L7LU10 A0A2P8YX69 A0A1B6KR03 A0A1B6DNL3 A0A1B6EHT1 A0A1B6IPS5 A0A2J7QLR3 E0W0A3 K7IWW7 A0A2S2PG70 A0A232EZP2 A0A182PFG1 A0A067RT58 E2BNI3 A0A182JML1 F4WR50 A0A182QQC1 W5JJE7 A0A151WEW2 A0A182FAA2 A0A084W4Y8 A0A182TYE4 A0A182XVL5 A0A195FKI4 E2AP81 A0A0L7QKP1 Q7PQT3 A0A182XEA8 A0A182N6Q7 A0A182V632 A0A182HJ12 A0A195BAS2 A0A158NU72 A0A151IQE1 A0A182K7P2 A0A1A9WWV3 A0A182RYE1 E9IHJ4 A0A1W7R8C1 A0A0K8VE71 A0A1B0EXN1 A0A182MGH5 A0A026WAF1 A0A1I8NNZ5 A0A154PF94 A0A1I8N754 A0A182W2U9 B3M2V1 B4LZ77 B0X9F7 A0A0M4EQU5 Q293U6 B4GLM0 A0A3B0JMJ9 B4KCC3 B4NB43 B4JUH7 W8BQE5 B3NZ98 A0A088AFG5 A0A1B6CFM7 Q9VEB7 B4IB91 A0A1W4U8E9 A0A310S7F4 A0A2A3EC89 V9IED8 B4PL80 A0A336L3U0 X1XCH0 A0A0K8U9F6 A0A182LC64 B4QUI4 A0A1B0D7X8 A0A2J7QMA2 Q17DW7 A0A023ERZ5 A0A182GTK0 Q7QBD5 A0A182HJA1 A0A182XE21 A0A1I8JUR4 A0A3F2Z0C7 A0A182S5M1 A0A2A4JMI3 A0A1Q3FS08 W8BSE4 A0A182K9D3 A0A182N5A7 A0A182PE46
Pubmed
19121390
28756777
26354079
22118469
26227816
29403074
+ More
20566863 20075255 28648823 24845553 20798317 21719571 20920257 23761445 24438588 25244985 12364791 14747013 17210077 21347285 21282665 24508170 30249741 25315136 17994087 15632085 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 17510324 24945155 26483478
20566863 20075255 28648823 24845553 20798317 21719571 20920257 23761445 24438588 25244985 12364791 14747013 17210077 21347285 21282665 24508170 30249741 25315136 17994087 15632085 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 17510324 24945155 26483478
EMBL
BABH01039109
ODYU01002550
SOQ40202.1
NWSH01002258
PCG68784.1
KZ149948
+ More
PZC76715.1 RSAL01000001 RVE55236.1 KQ460367 KPJ15483.1 AGBW02013189 OWR43892.1 JTDY01000101 KOB78864.1 PYGN01000305 PSN48850.1 GEBQ01026117 JAT13860.1 GEDC01010034 JAS27264.1 GECZ01032284 JAS37485.1 GECU01018803 JAS88903.1 NEVH01013241 PNF29531.1 DS235857 EEB19059.1 AAZX01003786 GGMR01015810 MBY28429.1 NNAY01001437 OXU23976.1 KK852478 KDR23004.1 GL449414 EFN82762.1 GL888284 EGI63277.1 AXCN02000061 ADMH02001318 ETN63025.1 KQ983238 KYQ46352.1 ATLV01020424 KE525302 KFB45282.1 KQ981512 KYN40866.1 GL441481 EFN64754.1 KQ414940 KOC59177.1 AAAB01008879 EAA08343.5 APCN01002114 KQ976532 KYM81646.1 ADTU01026281 KQ976780 KYN08371.1 GL763289 EFZ19936.1 GEHC01000247 JAV47398.1 GDHF01015619 GDHF01015100 GDHF01005326 JAI36695.1 JAI37214.1 JAI46988.1 AJWK01013735 AJWK01013736 AJWK01013737 AJWK01013738 AJWK01013739 AJWK01013740 AXCM01000406 KK107323 QOIP01000002 EZA52601.1 RLU25568.1 KQ434892 KZC10556.1 CH902617 EDV43481.1 CH940650 EDW67084.1 DS232534 EDS43126.1 CP012526 ALC46772.1 CM000070 EAL29118.2 CH479185 EDW38444.1 OUUW01000007 SPP83455.1 CH933806 EDW13732.1 CH964232 EDW81007.1 CH916374 EDV91147.1 GAMC01011079 GAMC01011078 JAB95476.1 CH954181 EDV48640.1 GEDC01025001 JAS12297.1 AE014297 AY113202 AAF55510.2 AAM29207.1 CH480827 EDW44649.1 KQ777939 OAD52029.1 KZ288287 PBC29393.1 JR039381 AEY58664.1 CM000160 EDW95862.1 UFQS01001621 UFQT01001621 SSX11633.1 SSX31198.1 ABLF02038819 GDHF01029045 JAI23269.1 CM000364 EDX12484.1 AJVK01032945 NEVH01013215 PNF29697.1 CH477290 EAT44634.1 GAPW01001613 JAC11985.1 JXUM01087313 KQ563625 KXJ73581.1 EAA08488.3 APCN01002134 NWSH01001035 PCG72996.1 GFDL01004655 JAV30390.1 GAMC01010354 GAMC01010353 GAMC01010350 JAB96201.1
PZC76715.1 RSAL01000001 RVE55236.1 KQ460367 KPJ15483.1 AGBW02013189 OWR43892.1 JTDY01000101 KOB78864.1 PYGN01000305 PSN48850.1 GEBQ01026117 JAT13860.1 GEDC01010034 JAS27264.1 GECZ01032284 JAS37485.1 GECU01018803 JAS88903.1 NEVH01013241 PNF29531.1 DS235857 EEB19059.1 AAZX01003786 GGMR01015810 MBY28429.1 NNAY01001437 OXU23976.1 KK852478 KDR23004.1 GL449414 EFN82762.1 GL888284 EGI63277.1 AXCN02000061 ADMH02001318 ETN63025.1 KQ983238 KYQ46352.1 ATLV01020424 KE525302 KFB45282.1 KQ981512 KYN40866.1 GL441481 EFN64754.1 KQ414940 KOC59177.1 AAAB01008879 EAA08343.5 APCN01002114 KQ976532 KYM81646.1 ADTU01026281 KQ976780 KYN08371.1 GL763289 EFZ19936.1 GEHC01000247 JAV47398.1 GDHF01015619 GDHF01015100 GDHF01005326 JAI36695.1 JAI37214.1 JAI46988.1 AJWK01013735 AJWK01013736 AJWK01013737 AJWK01013738 AJWK01013739 AJWK01013740 AXCM01000406 KK107323 QOIP01000002 EZA52601.1 RLU25568.1 KQ434892 KZC10556.1 CH902617 EDV43481.1 CH940650 EDW67084.1 DS232534 EDS43126.1 CP012526 ALC46772.1 CM000070 EAL29118.2 CH479185 EDW38444.1 OUUW01000007 SPP83455.1 CH933806 EDW13732.1 CH964232 EDW81007.1 CH916374 EDV91147.1 GAMC01011079 GAMC01011078 JAB95476.1 CH954181 EDV48640.1 GEDC01025001 JAS12297.1 AE014297 AY113202 AAF55510.2 AAM29207.1 CH480827 EDW44649.1 KQ777939 OAD52029.1 KZ288287 PBC29393.1 JR039381 AEY58664.1 CM000160 EDW95862.1 UFQS01001621 UFQT01001621 SSX11633.1 SSX31198.1 ABLF02038819 GDHF01029045 JAI23269.1 CM000364 EDX12484.1 AJVK01032945 NEVH01013215 PNF29697.1 CH477290 EAT44634.1 GAPW01001613 JAC11985.1 JXUM01087313 KQ563625 KXJ73581.1 EAA08488.3 APCN01002134 NWSH01001035 PCG72996.1 GFDL01004655 JAV30390.1 GAMC01010354 GAMC01010353 GAMC01010350 JAB96201.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000037510
+ More
UP000245037 UP000235965 UP000009046 UP000002358 UP000215335 UP000075885 UP000027135 UP000008237 UP000075880 UP000007755 UP000075886 UP000000673 UP000075809 UP000069272 UP000030765 UP000075902 UP000076408 UP000078541 UP000000311 UP000053825 UP000007062 UP000076407 UP000075884 UP000075903 UP000075840 UP000078540 UP000005205 UP000078542 UP000075881 UP000091820 UP000075900 UP000092461 UP000075883 UP000053097 UP000279307 UP000095300 UP000076502 UP000095301 UP000075920 UP000007801 UP000008792 UP000002320 UP000092553 UP000001819 UP000008744 UP000268350 UP000009192 UP000007798 UP000001070 UP000008711 UP000005203 UP000000803 UP000001292 UP000192221 UP000242457 UP000002282 UP000007819 UP000075882 UP000000304 UP000092462 UP000008820 UP000069940 UP000249989 UP000075901
UP000245037 UP000235965 UP000009046 UP000002358 UP000215335 UP000075885 UP000027135 UP000008237 UP000075880 UP000007755 UP000075886 UP000000673 UP000075809 UP000069272 UP000030765 UP000075902 UP000076408 UP000078541 UP000000311 UP000053825 UP000007062 UP000076407 UP000075884 UP000075903 UP000075840 UP000078540 UP000005205 UP000078542 UP000075881 UP000091820 UP000075900 UP000092461 UP000075883 UP000053097 UP000279307 UP000095300 UP000076502 UP000095301 UP000075920 UP000007801 UP000008792 UP000002320 UP000092553 UP000001819 UP000008744 UP000268350 UP000009192 UP000007798 UP000001070 UP000008711 UP000005203 UP000000803 UP000001292 UP000192221 UP000242457 UP000002282 UP000007819 UP000075882 UP000000304 UP000092462 UP000008820 UP000069940 UP000249989 UP000075901
Pfam
PF02958 EcKinase
Interpro
ProteinModelPortal
H9JGK2
A0A2H1VII8
A0A2A4JB62
A0A2W1BSV6
A0A3S2PLX5
A0A194RCB3
+ More
A0A212EQW9 A0A0L7LU10 A0A2P8YX69 A0A1B6KR03 A0A1B6DNL3 A0A1B6EHT1 A0A1B6IPS5 A0A2J7QLR3 E0W0A3 K7IWW7 A0A2S2PG70 A0A232EZP2 A0A182PFG1 A0A067RT58 E2BNI3 A0A182JML1 F4WR50 A0A182QQC1 W5JJE7 A0A151WEW2 A0A182FAA2 A0A084W4Y8 A0A182TYE4 A0A182XVL5 A0A195FKI4 E2AP81 A0A0L7QKP1 Q7PQT3 A0A182XEA8 A0A182N6Q7 A0A182V632 A0A182HJ12 A0A195BAS2 A0A158NU72 A0A151IQE1 A0A182K7P2 A0A1A9WWV3 A0A182RYE1 E9IHJ4 A0A1W7R8C1 A0A0K8VE71 A0A1B0EXN1 A0A182MGH5 A0A026WAF1 A0A1I8NNZ5 A0A154PF94 A0A1I8N754 A0A182W2U9 B3M2V1 B4LZ77 B0X9F7 A0A0M4EQU5 Q293U6 B4GLM0 A0A3B0JMJ9 B4KCC3 B4NB43 B4JUH7 W8BQE5 B3NZ98 A0A088AFG5 A0A1B6CFM7 Q9VEB7 B4IB91 A0A1W4U8E9 A0A310S7F4 A0A2A3EC89 V9IED8 B4PL80 A0A336L3U0 X1XCH0 A0A0K8U9F6 A0A182LC64 B4QUI4 A0A1B0D7X8 A0A2J7QMA2 Q17DW7 A0A023ERZ5 A0A182GTK0 Q7QBD5 A0A182HJA1 A0A182XE21 A0A1I8JUR4 A0A3F2Z0C7 A0A182S5M1 A0A2A4JMI3 A0A1Q3FS08 W8BSE4 A0A182K9D3 A0A182N5A7 A0A182PE46
A0A212EQW9 A0A0L7LU10 A0A2P8YX69 A0A1B6KR03 A0A1B6DNL3 A0A1B6EHT1 A0A1B6IPS5 A0A2J7QLR3 E0W0A3 K7IWW7 A0A2S2PG70 A0A232EZP2 A0A182PFG1 A0A067RT58 E2BNI3 A0A182JML1 F4WR50 A0A182QQC1 W5JJE7 A0A151WEW2 A0A182FAA2 A0A084W4Y8 A0A182TYE4 A0A182XVL5 A0A195FKI4 E2AP81 A0A0L7QKP1 Q7PQT3 A0A182XEA8 A0A182N6Q7 A0A182V632 A0A182HJ12 A0A195BAS2 A0A158NU72 A0A151IQE1 A0A182K7P2 A0A1A9WWV3 A0A182RYE1 E9IHJ4 A0A1W7R8C1 A0A0K8VE71 A0A1B0EXN1 A0A182MGH5 A0A026WAF1 A0A1I8NNZ5 A0A154PF94 A0A1I8N754 A0A182W2U9 B3M2V1 B4LZ77 B0X9F7 A0A0M4EQU5 Q293U6 B4GLM0 A0A3B0JMJ9 B4KCC3 B4NB43 B4JUH7 W8BQE5 B3NZ98 A0A088AFG5 A0A1B6CFM7 Q9VEB7 B4IB91 A0A1W4U8E9 A0A310S7F4 A0A2A3EC89 V9IED8 B4PL80 A0A336L3U0 X1XCH0 A0A0K8U9F6 A0A182LC64 B4QUI4 A0A1B0D7X8 A0A2J7QMA2 Q17DW7 A0A023ERZ5 A0A182GTK0 Q7QBD5 A0A182HJA1 A0A182XE21 A0A1I8JUR4 A0A3F2Z0C7 A0A182S5M1 A0A2A4JMI3 A0A1Q3FS08 W8BSE4 A0A182K9D3 A0A182N5A7 A0A182PE46
Ontologies
PANTHER
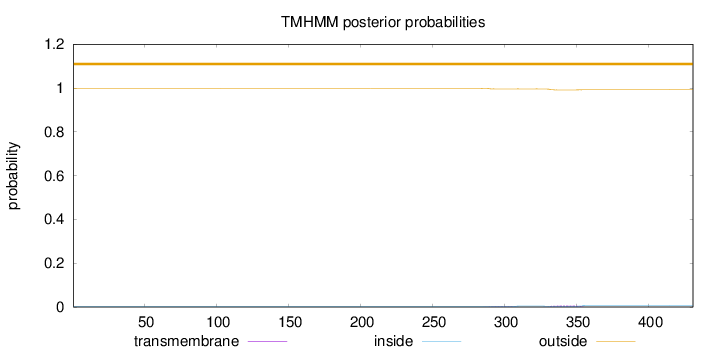
Topology
Length:
431
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17619
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00170
outside
1 - 431
Population Genetic Test Statistics
Pi
179.883238
Theta
138.791353
Tajima's D
1.187741
CLR
0.421729
CSRT
0.712414379281036
Interpretation
Uncertain
