Gene
KWMTBOMO04196
Pre Gene Modal
BGIBMGA008634
Annotation
PREDICTED:_inverted_formin-2-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.312 Nuclear Reliability : 1.691
Sequence
CDS
ATGTGTTCGTCAAGGGCCATAAAAAGATTGACTAAGTCTTGCGGTCACTTCAATTGGAATATATTATTTTTAGGTATTCCACCACCACCTGCACCGATGCTACCACCGCCTCCGCCGTGTGGACCGCCACCTCCCCCAGTCCCACCTCCCCTATCGGCTCCATCCGCTCCTATTCCACCAGCTCCAGTAATGGACCTAAAGCTGCCCCAACAAGAAACCCCTCTGCCTAAAACCAAAATGAAAACTATTAATTGGAATAAAATACCTAACAGTAAGGTGGTGGGACAGAACAACATCTGGTCAATAGTAGCGACAAGTCACAAACACTCACCGAAAACAGAGTTGGATTGGAACGAAATAGAAGATCTATTTTGTCAACAGATTCAACCGACCGGCTCGGCGGGGTCGTCGCCTCGCTTGGGCCGTAGCCCCATCTGCGACGCGTCCGGAGAAAAGAAACCGAGGAAAGAGTCCTCCGAGATCACGCTCCTCGACGGCAAGAGGAGCCTTAACGTCAACATATTCTTAAAGCAGTTCCGAAGTTCCAATGAAGATATTATACAGTTAATTAGAGAAGGTTCTCATGACGATATCGGCACGGAAAAGCTTCGTGGACTTTTGAAGATACTACCCGAAATAGACGAGTGCGAGATGCTTAAAGCGTTCTCCGGTGACGTCACAAAGCTCGGGAACGCGGAGAAGTTTCTGCTACAACTGATACAAGTACCCAATTACAAGTTACGTATAGAATGCATGCTGCTGAAAGAGGAATGGCCGTCCACTACTGGTTACTTGGAGAGCGCCATCAACGCAATCCTGGTTGCCGGAGATGACCTCATGTCTTCAAGAGCACTACAGGAGGTGCTGTATATAGCTTTAATAGCTGGTAATTTCCTCAACGCTGGTGGCTATGCGGGCGGCGCGGCCGGTGTTAAGCTGTCCTCACTTCAGAAGCTACAGGACATCAGGGCGAACAAGCCTGGAATCACTCTTATGCATTATGTGGCGCTGCAAGCAGAGCGCAAGAACAAAGAACTGACACATTTCGCGGATGATACGCGAGTTTTGGAAGAAGCTGCTAAGGCATCAGTCGAACAACTCCACAACGAGATCAAGACCCTAGAGACCAGGATCGGGGTGCTGAAGAAGGATATTTTGTTGCCAACCACCCAGCCTGATATTAGTCGGCAGATGGGAGAGTTTCTAAAGGTCGCTGAACAAGAAATATCAGTACTGAATAAAGACATGGAGGAAGTGGAAACCCTCCGAAAGCAGCTGGCGGAGTTCTTCTGCGAAGACTCCGTGTCCTTCAAACTTGAGGAATGCTTTAAGTTCCTCTGGTCCCAAGTCGCGTGGTGGTCGGTCTGA
Protein
MCSSRAIKRLTKSCGHFNWNILFLGIPPPPAPMLPPPPPCGPPPPPVPPPLSAPSAPIPPAPVMDLKLPQQETPLPKTKMKTINWNKIPNSKVVGQNNIWSIVATSHKHSPKTELDWNEIEDLFCQQIQPTGSAGSSPRLGRSPICDASGEKKPRKESSEITLLDGKRSLNVNIFLKQFRSSNEDIIQLIREGSHDDIGTEKLRGLLKILPEIDECEMLKAFSGDVTKLGNAEKFLLQLIQVPNYKLRIECMLLKEEWPSTTGYLESAINAILVAGDDLMSSRALQEVLYIALIAGNFLNAGGYAGGAAGVKLSSLQKLQDIRANKPGITLMHYVALQAERKNKELTHFADDTRVLEEAAKASVEQLHNEIKTLETRIGVLKKDILLPTTQPDISRQMGEFLKVAEQEISVLNKDMEEVETLRKQLAEFFCEDSVSFKLEECFKFLWSQVAWWSV
Summary
Uniprot
H9JGI7
A0A2W1BVP0
A0A2A4JBK6
A0A2H1VHH3
A0A194RDE2
A0A1E1W0K6
+ More
A0A194PW81 A0A3S2LH32 A0A212ER08 A0A2R7WVQ9 A0A2J7QQI7 A0A2J7QQH9 A0A1B6MSP7 A0A1B6D1V3 A0A0A9WV25 A0A0A9WLQ7 A0A1B6CF20 A0A067R850 A0A0A9WQB4 E0VWW6 A0A1B6FIR0 A0A1B6LXU9 W8BNN4 W8B8R1 A0A1Y1M1V1 A0A154P3G5 A0A034VWG5 A0A310SQD0 A0A232F420 K7J972 A0A0J7L2K1 X1WIB3 A0A0M9A7G1 A0A088A383 A0A0C9RE13 A0A026WS92 E2AC02 A0A146M1M5 T1IAV0 A0A1J1IDB3 A0A0K8V0R2 A0A0A1WPS2 A0A224XGJ7 W4VR99 A0A1Y1M246 A0A0L7QXG6 A0A0L7L339 A0A3B0JYR5 A0A1W4XHH1 E2B6K2 Q86C16 A0A0R1DW96 B3NFI4 A0A2M3ZGM2 A8JNM1 B4PK24 A0A1S4FPJ9 A0A1W4UGM1 A0A3B0J455 Q16U10 B0WJ07 A0A182GFN0 A0A139WDE5 D6X509 B4GR44 B5DPB2 A0A2P8ZFQ9 A0A195BH58 A0A158NL62 A0A139WAN0 A0A195F1T9 A0A182QQ11 A0A2M3Z0J1 A0A151JSF7 A0A0P9C1Z7 A0A2M4B8P5 B3M7X7 A0A2M4B8N9 A0A2M4AAP3 A0A2M4AAV4 A0A182VQZ9 A0A182IRU6 A0A182JXA0 A0A182FQI8 A0A182PC42 A0A182LSN7 A0A182YJS2 A0A182R1K6 A0A0J9RRT5 A0A182N5Z4 B4MMM5 A0A182V3K9 A0A182TNC8 B4HV19 W5J284 A0A2M4CG00 A0A2K6VB98 A0A2M4CIH1 A0A182I0D6 Q7PYF6
A0A194PW81 A0A3S2LH32 A0A212ER08 A0A2R7WVQ9 A0A2J7QQI7 A0A2J7QQH9 A0A1B6MSP7 A0A1B6D1V3 A0A0A9WV25 A0A0A9WLQ7 A0A1B6CF20 A0A067R850 A0A0A9WQB4 E0VWW6 A0A1B6FIR0 A0A1B6LXU9 W8BNN4 W8B8R1 A0A1Y1M1V1 A0A154P3G5 A0A034VWG5 A0A310SQD0 A0A232F420 K7J972 A0A0J7L2K1 X1WIB3 A0A0M9A7G1 A0A088A383 A0A0C9RE13 A0A026WS92 E2AC02 A0A146M1M5 T1IAV0 A0A1J1IDB3 A0A0K8V0R2 A0A0A1WPS2 A0A224XGJ7 W4VR99 A0A1Y1M246 A0A0L7QXG6 A0A0L7L339 A0A3B0JYR5 A0A1W4XHH1 E2B6K2 Q86C16 A0A0R1DW96 B3NFI4 A0A2M3ZGM2 A8JNM1 B4PK24 A0A1S4FPJ9 A0A1W4UGM1 A0A3B0J455 Q16U10 B0WJ07 A0A182GFN0 A0A139WDE5 D6X509 B4GR44 B5DPB2 A0A2P8ZFQ9 A0A195BH58 A0A158NL62 A0A139WAN0 A0A195F1T9 A0A182QQ11 A0A2M3Z0J1 A0A151JSF7 A0A0P9C1Z7 A0A2M4B8P5 B3M7X7 A0A2M4B8N9 A0A2M4AAP3 A0A2M4AAV4 A0A182VQZ9 A0A182IRU6 A0A182JXA0 A0A182FQI8 A0A182PC42 A0A182LSN7 A0A182YJS2 A0A182R1K6 A0A0J9RRT5 A0A182N5Z4 B4MMM5 A0A182V3K9 A0A182TNC8 B4HV19 W5J284 A0A2M4CG00 A0A2K6VB98 A0A2M4CIH1 A0A182I0D6 Q7PYF6
Pubmed
19121390
28756777
26354079
22118469
25401762
24845553
+ More
20566863 24495485 28004739 25348373 28648823 20075255 24508170 20798317 26823975 25830018 26227816 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 17510324 26483478 18362917 19820115 15632085 29403074 21347285 25244985 22936249 20920257 12364791
20566863 24495485 28004739 25348373 28648823 20075255 24508170 20798317 26823975 25830018 26227816 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 17510324 26483478 18362917 19820115 15632085 29403074 21347285 25244985 22936249 20920257 12364791
EMBL
BABH01041281
BABH01041282
KZ149948
PZC76716.1
NWSH01002258
PCG68783.1
+ More
ODYU01002550 SOQ40201.1 KQ460367 KPJ15484.1 GDQN01010595 JAT80459.1 KQ459589 KPI97636.1 RSAL01000126 RVE46552.1 AGBW02013189 OWR43891.1 KK855716 PTY23634.1 NEVH01012087 PNF30844.1 PNF30845.1 GEBQ01001023 JAT38954.1 GEDC01017648 JAS19650.1 GBHO01034919 JAG08685.1 GBHO01034920 JAG08684.1 GEDC01025242 JAS12056.1 KK852818 KDR15721.1 GBHO01034921 JAG08683.1 DS235824 EEB17872.1 GECZ01019709 JAS50060.1 GEBQ01011441 JAT28536.1 GAMC01011549 JAB95006.1 GAMC01011548 JAB95007.1 GEZM01042514 JAV79804.1 KQ434809 KZC06411.1 GAKP01012782 JAC46170.1 KQ761039 OAD58367.1 NNAY01001080 OXU25218.1 AAZX01002215 LBMM01001105 KMQ96916.1 ABLF02036636 ABLF02036637 KQ435719 KOX78790.1 GBYB01005206 JAG74973.1 KK107111 EZA58897.1 GL438389 EFN69020.1 GDHC01006044 GDHC01004918 JAQ12585.1 JAQ13711.1 ACPB03005776 ACPB03005777 CVRI01000045 CRK96966.1 GDHF01019797 JAI32517.1 GBXI01013879 GBXI01001749 JAD00413.1 JAD12543.1 GFTR01008846 JAW07580.1 GANO01003806 JAB56065.1 GEZM01042515 JAV79803.1 KQ414704 KOC63312.1 JTDY01003226 KOB69923.1 OUUW01000002 SPP76248.1 GL445975 EFN88694.1 AE014296 AB061681 AAX52757.1 BAC76439.1 CM000159 KRK01321.1 CH954178 EDV50526.2 GGFM01006908 MBW27659.1 ABW08480.1 EDW93704.2 SPP76247.1 CH477635 EAT38008.1 DS231954 EDS28868.1 JXUM01059985 JXUM01059986 JXUM01059987 JXUM01059988 KQ562081 KXJ76733.1 KQ971361 KYB25875.1 KQ971381 EEZ97607.1 CH479188 EDW40229.1 CH379069 EDY73451.1 PYGN01000072 PSN55318.1 KQ976488 KYM83506.1 ADTU01019263 ADTU01019264 ADTU01019265 ADTU01019266 KYB24978.1 KQ981866 KYN34064.1 AXCN02000191 GGFM01001289 MBW22040.1 KQ978530 KYN30303.1 CH902618 KPU77675.1 GGFJ01000273 MBW49414.1 EDV38850.2 GGFJ01000221 MBW49362.1 GGFK01004540 MBW37861.1 GGFK01004521 MBW37842.1 AXCM01003486 CM002912 KMY98019.1 CH963847 EDW73431.2 CH480817 EDW50790.1 ADMH02002134 ETN58367.1 GGFL01000094 MBW64272.1 GGFL01000849 MBW65027.1 GGFL01000857 MBW65035.1 APCN01002070 AAAB01008987 EAA01017.5
ODYU01002550 SOQ40201.1 KQ460367 KPJ15484.1 GDQN01010595 JAT80459.1 KQ459589 KPI97636.1 RSAL01000126 RVE46552.1 AGBW02013189 OWR43891.1 KK855716 PTY23634.1 NEVH01012087 PNF30844.1 PNF30845.1 GEBQ01001023 JAT38954.1 GEDC01017648 JAS19650.1 GBHO01034919 JAG08685.1 GBHO01034920 JAG08684.1 GEDC01025242 JAS12056.1 KK852818 KDR15721.1 GBHO01034921 JAG08683.1 DS235824 EEB17872.1 GECZ01019709 JAS50060.1 GEBQ01011441 JAT28536.1 GAMC01011549 JAB95006.1 GAMC01011548 JAB95007.1 GEZM01042514 JAV79804.1 KQ434809 KZC06411.1 GAKP01012782 JAC46170.1 KQ761039 OAD58367.1 NNAY01001080 OXU25218.1 AAZX01002215 LBMM01001105 KMQ96916.1 ABLF02036636 ABLF02036637 KQ435719 KOX78790.1 GBYB01005206 JAG74973.1 KK107111 EZA58897.1 GL438389 EFN69020.1 GDHC01006044 GDHC01004918 JAQ12585.1 JAQ13711.1 ACPB03005776 ACPB03005777 CVRI01000045 CRK96966.1 GDHF01019797 JAI32517.1 GBXI01013879 GBXI01001749 JAD00413.1 JAD12543.1 GFTR01008846 JAW07580.1 GANO01003806 JAB56065.1 GEZM01042515 JAV79803.1 KQ414704 KOC63312.1 JTDY01003226 KOB69923.1 OUUW01000002 SPP76248.1 GL445975 EFN88694.1 AE014296 AB061681 AAX52757.1 BAC76439.1 CM000159 KRK01321.1 CH954178 EDV50526.2 GGFM01006908 MBW27659.1 ABW08480.1 EDW93704.2 SPP76247.1 CH477635 EAT38008.1 DS231954 EDS28868.1 JXUM01059985 JXUM01059986 JXUM01059987 JXUM01059988 KQ562081 KXJ76733.1 KQ971361 KYB25875.1 KQ971381 EEZ97607.1 CH479188 EDW40229.1 CH379069 EDY73451.1 PYGN01000072 PSN55318.1 KQ976488 KYM83506.1 ADTU01019263 ADTU01019264 ADTU01019265 ADTU01019266 KYB24978.1 KQ981866 KYN34064.1 AXCN02000191 GGFM01001289 MBW22040.1 KQ978530 KYN30303.1 CH902618 KPU77675.1 GGFJ01000273 MBW49414.1 EDV38850.2 GGFJ01000221 MBW49362.1 GGFK01004540 MBW37861.1 GGFK01004521 MBW37842.1 AXCM01003486 CM002912 KMY98019.1 CH963847 EDW73431.2 CH480817 EDW50790.1 ADMH02002134 ETN58367.1 GGFL01000094 MBW64272.1 GGFL01000849 MBW65027.1 GGFL01000857 MBW65035.1 APCN01002070 AAAB01008987 EAA01017.5
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000235965 UP000027135 UP000009046 UP000076502 UP000215335 UP000002358 UP000036403 UP000007819 UP000053105 UP000005203 UP000053097 UP000000311 UP000015103 UP000183832 UP000053825 UP000037510 UP000268350 UP000192223 UP000008237 UP000000803 UP000002282 UP000008711 UP000192221 UP000008820 UP000002320 UP000069940 UP000249989 UP000007266 UP000008744 UP000001819 UP000245037 UP000078540 UP000005205 UP000078541 UP000075886 UP000078492 UP000007801 UP000075920 UP000075880 UP000075881 UP000069272 UP000075885 UP000075883 UP000076408 UP000075900 UP000075884 UP000007798 UP000075903 UP000075902 UP000001292 UP000000673 UP000075840 UP000007062
UP000235965 UP000027135 UP000009046 UP000076502 UP000215335 UP000002358 UP000036403 UP000007819 UP000053105 UP000005203 UP000053097 UP000000311 UP000015103 UP000183832 UP000053825 UP000037510 UP000268350 UP000192223 UP000008237 UP000000803 UP000002282 UP000008711 UP000192221 UP000008820 UP000002320 UP000069940 UP000249989 UP000007266 UP000008744 UP000001819 UP000245037 UP000078540 UP000005205 UP000078541 UP000075886 UP000078492 UP000007801 UP000075920 UP000075880 UP000075881 UP000069272 UP000075885 UP000075883 UP000076408 UP000075900 UP000075884 UP000007798 UP000075903 UP000075902 UP000001292 UP000000673 UP000075840 UP000007062
Interpro
Gene 3D
ProteinModelPortal
H9JGI7
A0A2W1BVP0
A0A2A4JBK6
A0A2H1VHH3
A0A194RDE2
A0A1E1W0K6
+ More
A0A194PW81 A0A3S2LH32 A0A212ER08 A0A2R7WVQ9 A0A2J7QQI7 A0A2J7QQH9 A0A1B6MSP7 A0A1B6D1V3 A0A0A9WV25 A0A0A9WLQ7 A0A1B6CF20 A0A067R850 A0A0A9WQB4 E0VWW6 A0A1B6FIR0 A0A1B6LXU9 W8BNN4 W8B8R1 A0A1Y1M1V1 A0A154P3G5 A0A034VWG5 A0A310SQD0 A0A232F420 K7J972 A0A0J7L2K1 X1WIB3 A0A0M9A7G1 A0A088A383 A0A0C9RE13 A0A026WS92 E2AC02 A0A146M1M5 T1IAV0 A0A1J1IDB3 A0A0K8V0R2 A0A0A1WPS2 A0A224XGJ7 W4VR99 A0A1Y1M246 A0A0L7QXG6 A0A0L7L339 A0A3B0JYR5 A0A1W4XHH1 E2B6K2 Q86C16 A0A0R1DW96 B3NFI4 A0A2M3ZGM2 A8JNM1 B4PK24 A0A1S4FPJ9 A0A1W4UGM1 A0A3B0J455 Q16U10 B0WJ07 A0A182GFN0 A0A139WDE5 D6X509 B4GR44 B5DPB2 A0A2P8ZFQ9 A0A195BH58 A0A158NL62 A0A139WAN0 A0A195F1T9 A0A182QQ11 A0A2M3Z0J1 A0A151JSF7 A0A0P9C1Z7 A0A2M4B8P5 B3M7X7 A0A2M4B8N9 A0A2M4AAP3 A0A2M4AAV4 A0A182VQZ9 A0A182IRU6 A0A182JXA0 A0A182FQI8 A0A182PC42 A0A182LSN7 A0A182YJS2 A0A182R1K6 A0A0J9RRT5 A0A182N5Z4 B4MMM5 A0A182V3K9 A0A182TNC8 B4HV19 W5J284 A0A2M4CG00 A0A2K6VB98 A0A2M4CIH1 A0A182I0D6 Q7PYF6
A0A194PW81 A0A3S2LH32 A0A212ER08 A0A2R7WVQ9 A0A2J7QQI7 A0A2J7QQH9 A0A1B6MSP7 A0A1B6D1V3 A0A0A9WV25 A0A0A9WLQ7 A0A1B6CF20 A0A067R850 A0A0A9WQB4 E0VWW6 A0A1B6FIR0 A0A1B6LXU9 W8BNN4 W8B8R1 A0A1Y1M1V1 A0A154P3G5 A0A034VWG5 A0A310SQD0 A0A232F420 K7J972 A0A0J7L2K1 X1WIB3 A0A0M9A7G1 A0A088A383 A0A0C9RE13 A0A026WS92 E2AC02 A0A146M1M5 T1IAV0 A0A1J1IDB3 A0A0K8V0R2 A0A0A1WPS2 A0A224XGJ7 W4VR99 A0A1Y1M246 A0A0L7QXG6 A0A0L7L339 A0A3B0JYR5 A0A1W4XHH1 E2B6K2 Q86C16 A0A0R1DW96 B3NFI4 A0A2M3ZGM2 A8JNM1 B4PK24 A0A1S4FPJ9 A0A1W4UGM1 A0A3B0J455 Q16U10 B0WJ07 A0A182GFN0 A0A139WDE5 D6X509 B4GR44 B5DPB2 A0A2P8ZFQ9 A0A195BH58 A0A158NL62 A0A139WAN0 A0A195F1T9 A0A182QQ11 A0A2M3Z0J1 A0A151JSF7 A0A0P9C1Z7 A0A2M4B8P5 B3M7X7 A0A2M4B8N9 A0A2M4AAP3 A0A2M4AAV4 A0A182VQZ9 A0A182IRU6 A0A182JXA0 A0A182FQI8 A0A182PC42 A0A182LSN7 A0A182YJS2 A0A182R1K6 A0A0J9RRT5 A0A182N5Z4 B4MMM5 A0A182V3K9 A0A182TNC8 B4HV19 W5J284 A0A2M4CG00 A0A2K6VB98 A0A2M4CIH1 A0A182I0D6 Q7PYF6
PDB
2Z6E
E-value=8.03417e-28,
Score=308
Ontologies
PANTHER
Topology
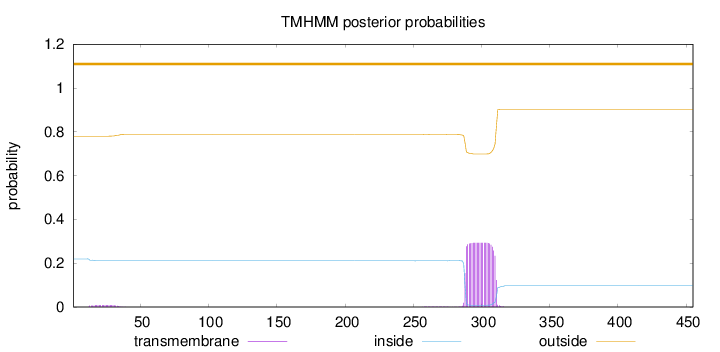
Length:
455
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.83531
Exp number, first 60 AAs:
0.1743
Total prob of N-in:
0.21976
outside
1 - 455
Population Genetic Test Statistics
Pi
234.569957
Theta
171.423495
Tajima's D
1.15873
CLR
0.019582
CSRT
0.706864656767162
Interpretation
Uncertain
