Gene
KWMTBOMO04192
Annotation
PREDICTED:_organic_cation_transporter_protein-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.968
Sequence
CDS
ATGGTGCAAAGATTCACCGTCGAAGTACCACAGAGCGCTAAAAAAGAAGACAAATTTGTCAGCTTAGTTGGAGAATTCGGAAGATGGCAGTTTTTAATTTTCAGCACCATATCTTTGGTGAAATTCTCATCGGGCACCGTCCAAATGGCTATACTCTTCTTGACCCCTAACTTGGTCTTCTGGTGTGAGAAGTTCGCAAATAATTCTAGAATAGTTCAGAATAACACTTGTTACAAGGATTGCGTAAAATACAGTTATGACACTAGCCCGTTTGAGGAGACTATCGTATCGGAATGGGACCTGGTCTGTGAGAGGAGATGGCTGGCGAGCTTCACACAGATGCTTCTACAATTTGGCATCTTAATTGGCAGTATCATATTTGGATTCTTTTCTGATAGATACGGTCGCAAAGTCACCTTCTTGATAGCGATTGCTGCAATGATTTTAATCGGTTTCGGGATCCCATTCGCGCCTGGATACACAACCTTCACTGTACTAAGATTCCTTCTGGGAGTCGCTACAGCTGGGACCATGGTCGTATCTTTCGTAATCATTATGGAGTCGATAGGCAATAAGTACAGAGAGGTCGTGGGCTGCTTATTTCAAATACCTTTCATTATAGGACACATATCAGTACCCCTCTTCGCGTATTACTTCCGAAGATGGAACTCCTACACACTCGCGCTTGCAGTACCTCCTCTCATATACCTGGGTTATTTCTTTCTGCTTTCTGAATCTCCGAGGTGGCTGGTCAGCGTAGGAAAAGTAGAAGAGGCCACGAGAATCGTTAAGAAGGCCGCTCTGATAAACAATCTGCCAACGGCGAAAATAGAGGACACACTAAAAAAGCTTGCCGAAGAAATCACCTCGAGCGAAGTCCAAAAGCCAAATTACTCTGATTTGTTTCGGAAGCCGTTGCTGGTGCAGTCGGTGTCCTGCTGTGCTATCTGGCTCGTCACGGGGGTCACGTTCTTCGGGTTCAACCAGTACATCAGCCAGTCAAGTCCAGACCCCTTTGTTAGCGTCGCAGCCGCTGGTGCTATTCAGATACCGTCCAACTTAATTTCAATTTGGCTGATCCGAAAACTAGGACGTAAAGTCACAACAACTATGTTCTCGGCCTTGGGAGGTCTCTGCATCATCACCCTAGGATGTGTCCCTGATATTTTCTGGCTGACCTTGACCCTGGGAACGTTGGGAGTCAGCTGCACTGCCATTATCGCGGCCACAATCTATATTTACACTTCAGAATTATATCCTACAGTCGTTAGGAACATGGCCTTGGGAGCTTGTTCCACCTCCATGAGGGTAGGGTCGATGTTGGCACCGTTCGTATCGAATCTATCAGTGACTGCGCCGTGGCTGCCGACCCTCGTCTTTGGCGTGGCACCCCTTTGTTCAGCGTTGGTTTGTCTGTTGCTGCCGGAGACGAAGGGGAAGACATTACTGAATAGTTTAAGGGAAGTAAACAATGGAAGATGA
Protein
MVQRFTVEVPQSAKKEDKFVSLVGEFGRWQFLIFSTISLVKFSSGTVQMAILFLTPNLVFWCEKFANNSRIVQNNTCYKDCVKYSYDTSPFEETIVSEWDLVCERRWLASFTQMLLQFGILIGSIIFGFFSDRYGRKVTFLIAIAAMILIGFGIPFAPGYTTFTVLRFLLGVATAGTMVVSFVIIMESIGNKYREVVGCLFQIPFIIGHISVPLFAYYFRRWNSYTLALAVPPLIYLGYFFLLSESPRWLVSVGKVEEATRIVKKAALINNLPTAKIEDTLKKLAEEITSSEVQKPNYSDLFRKPLLVQSVSCCAIWLVTGVTFFGFNQYISQSSPDPFVSVAAAGAIQIPSNLISIWLIRKLGRKVTTTMFSALGGLCIITLGCVPDIFWLTLTLGTLGVSCTAIIAATIYIYTSELYPTVVRNMALGACSTSMRVGSMLAPFVSNLSVTAPWLPTLVFGVAPLCSALVCLLLPETKGKTLLNSLREVNNGR
Summary
Uniprot
A0A2A4J3B3
A0A2W1BP27
A0A194RHR5
A0A212F8L3
A0A0L7LUK1
A0A194PY14
+ More
A0A2H1WBB8 I4DNF7 A0A194PMC3 A0A194QN55 H9JNB3 A0A2H1VGH7 A0A2H1VMD7 A0A2W1BFH6 A0A2A4K5L0 A0A2A4KA95 A0A2A4K9Y1 H9ISF5 A0A2H1W3H1 A0A1B0C9J9 A0A2A4K9Y5 V5IAH1 A0A1W4WYZ1 A0A1W4X9I8 A0A182H4N7 E1ZZY2 D6WKJ1 D6WKJ0 A0A023EUG3 Q17CS1 A0A1Q3FH65 A0A232EL12 D6X4T8 A0A088A3V4 B0X4Q4 A0A310SUT5 A0A158NPS8 A0A2W1BCH7 H9JNI6 K7JAK3 A0A151IP62 K7ITG9 A0A195ATZ5 A0A0C9PLB6 A0A182NFM5 U4UII8 A0A182K794 E9IX20 A0A182TEQ3 A0A194QK42 A0A151JT16 A0A195ELY0 A0A212EW65 A0A2A3ELA5 Q7QDB3 A0A182HUA7 A0A182WRH0 A0A1Y1KD57 A0A182VI17 A0A182PAB8 A0A1Y1K9J5 A0A182W090 A0A182Y4D6 A0A182MBN7 A0A154NXF3 A0A182QLT3 A0A194QIQ7 A0A0A1WZR4 A0A182RNN4 A0A0L7LUI9 N6T164 F4W7G9 A0A0K8UVC6 A0A151X8L1 A0A194PL20 A0A2S2QUM7 A0A2H1VUP3 E2BIH0 A0A084VXP9 A0A1J1IHN1 A0A034WDR5 A0A182F7G4 A0A2M4CTR9 A0A232F564 A0A2M4CU13 A0A195DRF1 A0A2J7RBY0 E9J9G6 A0A0L7R034 A0A195C6E8 A0A182JCT1 A0A1B0CZU5 A0A194PLC9 A0A195FCZ0 A0A0T6BBT7 A0A2H8TVD0 X1WIK7 A0A212FFS4 A0A3S2LHN8 Q28YE2
A0A2H1WBB8 I4DNF7 A0A194PMC3 A0A194QN55 H9JNB3 A0A2H1VGH7 A0A2H1VMD7 A0A2W1BFH6 A0A2A4K5L0 A0A2A4KA95 A0A2A4K9Y1 H9ISF5 A0A2H1W3H1 A0A1B0C9J9 A0A2A4K9Y5 V5IAH1 A0A1W4WYZ1 A0A1W4X9I8 A0A182H4N7 E1ZZY2 D6WKJ1 D6WKJ0 A0A023EUG3 Q17CS1 A0A1Q3FH65 A0A232EL12 D6X4T8 A0A088A3V4 B0X4Q4 A0A310SUT5 A0A158NPS8 A0A2W1BCH7 H9JNI6 K7JAK3 A0A151IP62 K7ITG9 A0A195ATZ5 A0A0C9PLB6 A0A182NFM5 U4UII8 A0A182K794 E9IX20 A0A182TEQ3 A0A194QK42 A0A151JT16 A0A195ELY0 A0A212EW65 A0A2A3ELA5 Q7QDB3 A0A182HUA7 A0A182WRH0 A0A1Y1KD57 A0A182VI17 A0A182PAB8 A0A1Y1K9J5 A0A182W090 A0A182Y4D6 A0A182MBN7 A0A154NXF3 A0A182QLT3 A0A194QIQ7 A0A0A1WZR4 A0A182RNN4 A0A0L7LUI9 N6T164 F4W7G9 A0A0K8UVC6 A0A151X8L1 A0A194PL20 A0A2S2QUM7 A0A2H1VUP3 E2BIH0 A0A084VXP9 A0A1J1IHN1 A0A034WDR5 A0A182F7G4 A0A2M4CTR9 A0A232F564 A0A2M4CU13 A0A195DRF1 A0A2J7RBY0 E9J9G6 A0A0L7R034 A0A195C6E8 A0A182JCT1 A0A1B0CZU5 A0A194PLC9 A0A195FCZ0 A0A0T6BBT7 A0A2H8TVD0 X1WIK7 A0A212FFS4 A0A3S2LHN8 Q28YE2
Pubmed
EMBL
NWSH01003717
PCG65892.1
KZ149948
PZC76718.1
KQ460367
KPJ15486.1
+ More
AGBW02009713 OWR50049.1 JTDY01000054 KOB79137.1 KQ459589 KPI97639.1 ODYU01007212 SOQ49794.1 AK402884 BAM19447.1 KQ459600 KPI94138.1 KQ461195 KPJ06789.1 BABH01019600 ODYU01002452 SOQ39959.1 ODYU01003353 SOQ42005.1 KZ150304 PZC71520.1 NWSH01000158 PCG78950.1 NWSH01000020 PCG80713.1 PCG80714.1 BABH01044534 BABH01044535 ODYU01006026 SOQ47506.1 AJWK01002434 AJWK01002435 AJWK01002436 PCG80708.1 GALX01001195 JAB67271.1 JXUM01109993 KQ565372 KXJ71040.1 GL435531 EFN73254.1 KQ971342 EFA04007.2 EFA04006.1 GAPW01001011 JAC12587.1 CH477305 EAT44124.1 GFDL01008134 JAV26911.1 NNAY01003667 OXU19035.1 KQ971381 EEZ97586.2 DS232350 EDS40471.1 KQ760085 OAD61975.1 ADTU01022726 KZ150301 PZC71534.1 BABH01019597 AAZX01002446 KQ976914 KYN07128.1 AAZX01006832 KQ976745 KYM75454.1 GBYB01001833 JAG71600.1 KB632314 ERL92208.1 GL766616 EFZ14896.1 KQ461198 KPJ05943.1 KQ982014 KYN30660.1 KQ978730 KYN28932.1 AGBW02012077 OWR45719.1 KZ288225 PBC31939.1 AAAB01008859 EAA07935.5 APCN01000614 GEZM01088532 JAV58130.1 GEZM01088533 JAV58129.1 AXCM01003853 KQ434769 KZC03774.1 AXCN02000238 KQ459185 KPJ03336.1 GBXI01013722 GBXI01009990 JAD00570.1 JAD04302.1 KOB79138.1 APGK01048466 KB741110 ENN73834.1 GL887844 EGI69842.1 GDHF01021687 JAI30627.1 KQ982409 KYQ56702.1 KPI94131.1 GGMS01012221 MBY81424.1 ODYU01004551 SOQ44555.1 GL448504 EFN84558.1 ATLV01018145 KE525215 KFB42743.1 CVRI01000052 CRK99712.1 GAKP01006475 JAC52477.1 GGFL01004566 MBW68744.1 NNAY01000983 OXU25618.1 GGFL01004591 MBW68769.1 KQ980581 KYN15387.1 NEVH01005889 PNF38331.1 GL769313 EFZ10529.1 KQ414672 KOC64208.1 KQ978219 KYM96439.1 AJVK01009733 KPI94132.1 KQ981673 KYN38256.1 LJIG01002173 KRT84767.1 GFXV01006391 MBW18196.1 ABLF02040797 ABLF02040821 ABLF02040843 ABLF02040845 AGBW02008774 OWR52573.1 RSAL01000008 RVE54031.1 CM000071 EAL26023.2 KRT02771.1 KRT02772.1 KRT02773.1 KRT02774.1
AGBW02009713 OWR50049.1 JTDY01000054 KOB79137.1 KQ459589 KPI97639.1 ODYU01007212 SOQ49794.1 AK402884 BAM19447.1 KQ459600 KPI94138.1 KQ461195 KPJ06789.1 BABH01019600 ODYU01002452 SOQ39959.1 ODYU01003353 SOQ42005.1 KZ150304 PZC71520.1 NWSH01000158 PCG78950.1 NWSH01000020 PCG80713.1 PCG80714.1 BABH01044534 BABH01044535 ODYU01006026 SOQ47506.1 AJWK01002434 AJWK01002435 AJWK01002436 PCG80708.1 GALX01001195 JAB67271.1 JXUM01109993 KQ565372 KXJ71040.1 GL435531 EFN73254.1 KQ971342 EFA04007.2 EFA04006.1 GAPW01001011 JAC12587.1 CH477305 EAT44124.1 GFDL01008134 JAV26911.1 NNAY01003667 OXU19035.1 KQ971381 EEZ97586.2 DS232350 EDS40471.1 KQ760085 OAD61975.1 ADTU01022726 KZ150301 PZC71534.1 BABH01019597 AAZX01002446 KQ976914 KYN07128.1 AAZX01006832 KQ976745 KYM75454.1 GBYB01001833 JAG71600.1 KB632314 ERL92208.1 GL766616 EFZ14896.1 KQ461198 KPJ05943.1 KQ982014 KYN30660.1 KQ978730 KYN28932.1 AGBW02012077 OWR45719.1 KZ288225 PBC31939.1 AAAB01008859 EAA07935.5 APCN01000614 GEZM01088532 JAV58130.1 GEZM01088533 JAV58129.1 AXCM01003853 KQ434769 KZC03774.1 AXCN02000238 KQ459185 KPJ03336.1 GBXI01013722 GBXI01009990 JAD00570.1 JAD04302.1 KOB79138.1 APGK01048466 KB741110 ENN73834.1 GL887844 EGI69842.1 GDHF01021687 JAI30627.1 KQ982409 KYQ56702.1 KPI94131.1 GGMS01012221 MBY81424.1 ODYU01004551 SOQ44555.1 GL448504 EFN84558.1 ATLV01018145 KE525215 KFB42743.1 CVRI01000052 CRK99712.1 GAKP01006475 JAC52477.1 GGFL01004566 MBW68744.1 NNAY01000983 OXU25618.1 GGFL01004591 MBW68769.1 KQ980581 KYN15387.1 NEVH01005889 PNF38331.1 GL769313 EFZ10529.1 KQ414672 KOC64208.1 KQ978219 KYM96439.1 AJVK01009733 KPI94132.1 KQ981673 KYN38256.1 LJIG01002173 KRT84767.1 GFXV01006391 MBW18196.1 ABLF02040797 ABLF02040821 ABLF02040843 ABLF02040845 AGBW02008774 OWR52573.1 RSAL01000008 RVE54031.1 CM000071 EAL26023.2 KRT02771.1 KRT02772.1 KRT02773.1 KRT02774.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000037510
UP000053268
UP000005204
+ More
UP000092461 UP000192223 UP000069940 UP000249989 UP000000311 UP000007266 UP000008820 UP000215335 UP000005203 UP000002320 UP000005205 UP000002358 UP000078542 UP000078540 UP000075884 UP000030742 UP000075881 UP000075902 UP000078541 UP000078492 UP000242457 UP000007062 UP000075840 UP000076407 UP000075903 UP000075885 UP000075920 UP000076408 UP000075883 UP000076502 UP000075886 UP000075900 UP000019118 UP000007755 UP000075809 UP000008237 UP000030765 UP000183832 UP000069272 UP000235965 UP000053825 UP000075880 UP000092462 UP000007819 UP000283053 UP000001819
UP000092461 UP000192223 UP000069940 UP000249989 UP000000311 UP000007266 UP000008820 UP000215335 UP000005203 UP000002320 UP000005205 UP000002358 UP000078542 UP000078540 UP000075884 UP000030742 UP000075881 UP000075902 UP000078541 UP000078492 UP000242457 UP000007062 UP000075840 UP000076407 UP000075903 UP000075885 UP000075920 UP000076408 UP000075883 UP000076502 UP000075886 UP000075900 UP000019118 UP000007755 UP000075809 UP000008237 UP000030765 UP000183832 UP000069272 UP000235965 UP000053825 UP000075880 UP000092462 UP000007819 UP000283053 UP000001819
Pfam
Interpro
IPR005828
MFS_sugar_transport-like
+ More
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR005829 Sugar_transporter_CS
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR017986 WD40_repeat_dom
IPR024977 Apc4_WD40_dom
IPR022100 Mcl1_mid
IPR036322 WD40_repeat_dom_sf
IPR011041 Quinoprot_gluc/sorb_DH
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR032098 Acyltransf_C
IPR002123 Plipid/glycerol_acylTrfase
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR005829 Sugar_transporter_CS
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR017986 WD40_repeat_dom
IPR024977 Apc4_WD40_dom
IPR022100 Mcl1_mid
IPR036322 WD40_repeat_dom_sf
IPR011041 Quinoprot_gluc/sorb_DH
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR032098 Acyltransf_C
IPR002123 Plipid/glycerol_acylTrfase
Gene 3D
CDD
ProteinModelPortal
A0A2A4J3B3
A0A2W1BP27
A0A194RHR5
A0A212F8L3
A0A0L7LUK1
A0A194PY14
+ More
A0A2H1WBB8 I4DNF7 A0A194PMC3 A0A194QN55 H9JNB3 A0A2H1VGH7 A0A2H1VMD7 A0A2W1BFH6 A0A2A4K5L0 A0A2A4KA95 A0A2A4K9Y1 H9ISF5 A0A2H1W3H1 A0A1B0C9J9 A0A2A4K9Y5 V5IAH1 A0A1W4WYZ1 A0A1W4X9I8 A0A182H4N7 E1ZZY2 D6WKJ1 D6WKJ0 A0A023EUG3 Q17CS1 A0A1Q3FH65 A0A232EL12 D6X4T8 A0A088A3V4 B0X4Q4 A0A310SUT5 A0A158NPS8 A0A2W1BCH7 H9JNI6 K7JAK3 A0A151IP62 K7ITG9 A0A195ATZ5 A0A0C9PLB6 A0A182NFM5 U4UII8 A0A182K794 E9IX20 A0A182TEQ3 A0A194QK42 A0A151JT16 A0A195ELY0 A0A212EW65 A0A2A3ELA5 Q7QDB3 A0A182HUA7 A0A182WRH0 A0A1Y1KD57 A0A182VI17 A0A182PAB8 A0A1Y1K9J5 A0A182W090 A0A182Y4D6 A0A182MBN7 A0A154NXF3 A0A182QLT3 A0A194QIQ7 A0A0A1WZR4 A0A182RNN4 A0A0L7LUI9 N6T164 F4W7G9 A0A0K8UVC6 A0A151X8L1 A0A194PL20 A0A2S2QUM7 A0A2H1VUP3 E2BIH0 A0A084VXP9 A0A1J1IHN1 A0A034WDR5 A0A182F7G4 A0A2M4CTR9 A0A232F564 A0A2M4CU13 A0A195DRF1 A0A2J7RBY0 E9J9G6 A0A0L7R034 A0A195C6E8 A0A182JCT1 A0A1B0CZU5 A0A194PLC9 A0A195FCZ0 A0A0T6BBT7 A0A2H8TVD0 X1WIK7 A0A212FFS4 A0A3S2LHN8 Q28YE2
A0A2H1WBB8 I4DNF7 A0A194PMC3 A0A194QN55 H9JNB3 A0A2H1VGH7 A0A2H1VMD7 A0A2W1BFH6 A0A2A4K5L0 A0A2A4KA95 A0A2A4K9Y1 H9ISF5 A0A2H1W3H1 A0A1B0C9J9 A0A2A4K9Y5 V5IAH1 A0A1W4WYZ1 A0A1W4X9I8 A0A182H4N7 E1ZZY2 D6WKJ1 D6WKJ0 A0A023EUG3 Q17CS1 A0A1Q3FH65 A0A232EL12 D6X4T8 A0A088A3V4 B0X4Q4 A0A310SUT5 A0A158NPS8 A0A2W1BCH7 H9JNI6 K7JAK3 A0A151IP62 K7ITG9 A0A195ATZ5 A0A0C9PLB6 A0A182NFM5 U4UII8 A0A182K794 E9IX20 A0A182TEQ3 A0A194QK42 A0A151JT16 A0A195ELY0 A0A212EW65 A0A2A3ELA5 Q7QDB3 A0A182HUA7 A0A182WRH0 A0A1Y1KD57 A0A182VI17 A0A182PAB8 A0A1Y1K9J5 A0A182W090 A0A182Y4D6 A0A182MBN7 A0A154NXF3 A0A182QLT3 A0A194QIQ7 A0A0A1WZR4 A0A182RNN4 A0A0L7LUI9 N6T164 F4W7G9 A0A0K8UVC6 A0A151X8L1 A0A194PL20 A0A2S2QUM7 A0A2H1VUP3 E2BIH0 A0A084VXP9 A0A1J1IHN1 A0A034WDR5 A0A182F7G4 A0A2M4CTR9 A0A232F564 A0A2M4CU13 A0A195DRF1 A0A2J7RBY0 E9J9G6 A0A0L7R034 A0A195C6E8 A0A182JCT1 A0A1B0CZU5 A0A194PLC9 A0A195FCZ0 A0A0T6BBT7 A0A2H8TVD0 X1WIK7 A0A212FFS4 A0A3S2LHN8 Q28YE2
PDB
4LDS
E-value=1.25215e-10,
Score=161
Ontologies
GO
Topology
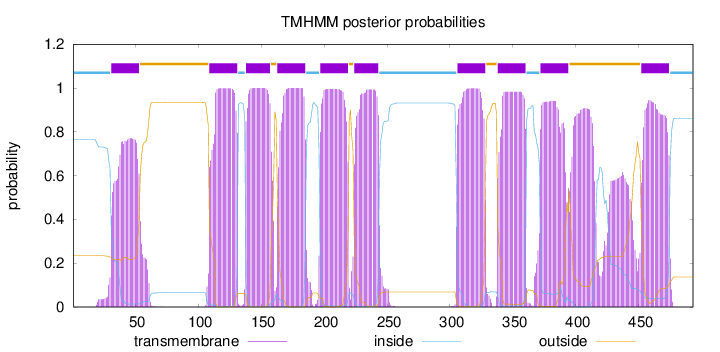
Length:
493
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
242.74011
Exp number, first 60 AAs:
18.08333
Total prob of N-in:
0.76555
POSSIBLE N-term signal
sequence
inside
1 - 30
TMhelix
31 - 53
outside
54 - 108
TMhelix
109 - 131
inside
132 - 137
TMhelix
138 - 157
outside
158 - 162
TMhelix
163 - 185
inside
186 - 196
TMhelix
197 - 219
outside
220 - 223
TMhelix
224 - 243
inside
244 - 305
TMhelix
306 - 328
outside
329 - 337
TMhelix
338 - 360
inside
361 - 371
TMhelix
372 - 394
outside
395 - 451
TMhelix
452 - 474
inside
475 - 493
Population Genetic Test Statistics
Pi
361.478913
Theta
194.349554
Tajima's D
4.00372
CLR
0.290085
CSRT
0.998650067496625
Interpretation
Uncertain
