Gene
KWMTBOMO04191
Pre Gene Modal
BGIBMGA008731
Annotation
PREDICTED:_pancreatic_triacylglycerol_lipase-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 0.953
Sequence
CDS
ATGTTTGTTTCAGTTCCGGGGGGAAGGTCGTTCACACCCAACGACATTATACTCCGGTACTATGGTAAGAATTCGACTGTGCCGACATCATATCGTTTTAGTGAGGTGAACAGACTTTTAGCTGACCCAACGTTTGATCCTAAGCGACCTACGGTACTATTTGCTCACGGGTATGTGGAGTTAGCGACCGATGAATCGGTACTAAGGATAGTTCGTGCCTACGTCCAGAGAGGCGGTTATAACATTCTAGTATTAGATTGGTCGAACATGGCCTTTGGAAACTACCTGCTTGTTTCGTTGGACGTACCTGTTACGGGCAAGAAAGCAGGCAAAGCACTGCTAAAGCTGTTAAAGGGGGGGCTGTCTTTGAAAGGGTTGCATTTAGTGGGACATTCACTCGGCGCGCATCTCTTGAGCTTTGCAGCAAAAGCTTTAGCAGCAAAAGGATTTACAGTACCCAGATTAACTGGTCTGGATCCCGCGTACCCAGGGTTCTACCCTCCGCTGCTGGCGGCGCACATGTCCAGCGAGGACGCTGACTTCGTGGACGTAATCCACACGGACGGGGGAGGTTTCGGTGCCCCCTCCAGCACCGGTCATGCCGACTTCTGGCCCAACGGTGGGCAGGCCAAGCAACCCGGCTGCCTGTCGTTCACTGTGCCTTTATCCAATGAAGATTTCTGCAGCCACTGGCGGTCGTGGGCCTTCTGGGCGGAGTCAGTCGAAGGGAGTCAGTTCATGAGTCGAAAGTGCCTAGATTATGACGCTTTTCTGCGCGGCCAGTGTCACGACGCACCCTCAGTGAGCATGGGGCTGGGAGCCACGTCTGAATTACGAGGGAATTTTTATCTTCGGACGGCCACTAGAACTCCGTATTCTCTAGGCATCAAAGGAGCAGAATAA
Protein
MFVSVPGGRSFTPNDIILRYYGKNSTVPTSYRFSEVNRLLADPTFDPKRPTVLFAHGYVELATDESVLRIVRAYVQRGGYNILVLDWSNMAFGNYLLVSLDVPVTGKKAGKALLKLLKGGLSLKGLHLVGHSLGAHLLSFAAKALAAKGFTVPRLTGLDPAYPGFYPPLLAAHMSSEDADFVDVIHTDGGGFGAPSSTGHADFWPNGGQAKQPGCLSFTVPLSNEDFCSHWRSWAFWAESVEGSQFMSRKCLDYDAFLRGQCHDAPSVSMGLGATSELRGNFYLRTATRTPYSLGIKGAE
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A2A4J1M9
A0A2W1B7Q2
A0A2H1W9L5
A0A194Q2K6
A0A194RCZ1
A0A3S2NUP7
+ More
B0W8V6 A0A182KUQ0 Q5TNF5 A0A182I4Y9 A0A1S4H040 A0A182VEG6 A0A1S4H0X8 A0A182TXT4 A0A182X2W4 A0A182SXZ4 A0A182WGZ2 A0A182YFE7 A0A182MGV1 U5EQP3 A0A336MI67 A0A1L8DJ03 A0A182QEE4 A0A182R9P9 A0A1B0CVI8 A0A2H1V975 A0A182PDD3 A0A2H1V1S5 A0A2M4BUY8 A0A2M3Z754 A0A2K6VB47 A0A182F5H0 A0A023EPI5 A0A023END9 A0A194R1U8 A0A194PH51 A0A2M4BU04 A0A182GL29 A0A182IUI2 A0A2M4AM81 A0A2M4AKZ7 A0A2M4AL49 A0A2M4CHM7 T1EAU4 H9JLK0 A0A336KHN4 A0A2M4CRR7 Q173Z1 Q173Z2 A0A336MGQ6 I4DNZ3 A0A1J1IX47 B4G838 Q29N87 A0A1B0D5A4 A0A3B0KIF6 A0A182NEW7 K7J451 A0A232F124 W8BCB3 A0A034VLB9 T1DGR4 J9HSR1 A0A194Q121 B4KE79 A0A1I8PDV7 A0A0N1PJG6 B4LS98 T1PIS7 A0A0A1XI69 A0A0A1WUY1 A0A1I8M5A8 A0A0K8UL76 B3MNS5 B4N0Y9 A0A0K8WAJ4 E2AIM4 A0A212EPM6 A0A232F8C9 A0A026WVQ0 A0A3L8DFK2 A0A1J1IQ95 B4JAK8 A0A2P8YQG3 D6WBV6 A0A158NQH7 K7IR20 A0A151X5R7 E9IDK7 A0A1W4X7R5 A0A0A1XE33 A0A1J1IPR1 A0A195CQD8 A0A1J0M5H9 J9JWA0
B0W8V6 A0A182KUQ0 Q5TNF5 A0A182I4Y9 A0A1S4H040 A0A182VEG6 A0A1S4H0X8 A0A182TXT4 A0A182X2W4 A0A182SXZ4 A0A182WGZ2 A0A182YFE7 A0A182MGV1 U5EQP3 A0A336MI67 A0A1L8DJ03 A0A182QEE4 A0A182R9P9 A0A1B0CVI8 A0A2H1V975 A0A182PDD3 A0A2H1V1S5 A0A2M4BUY8 A0A2M3Z754 A0A2K6VB47 A0A182F5H0 A0A023EPI5 A0A023END9 A0A194R1U8 A0A194PH51 A0A2M4BU04 A0A182GL29 A0A182IUI2 A0A2M4AM81 A0A2M4AKZ7 A0A2M4AL49 A0A2M4CHM7 T1EAU4 H9JLK0 A0A336KHN4 A0A2M4CRR7 Q173Z1 Q173Z2 A0A336MGQ6 I4DNZ3 A0A1J1IX47 B4G838 Q29N87 A0A1B0D5A4 A0A3B0KIF6 A0A182NEW7 K7J451 A0A232F124 W8BCB3 A0A034VLB9 T1DGR4 J9HSR1 A0A194Q121 B4KE79 A0A1I8PDV7 A0A0N1PJG6 B4LS98 T1PIS7 A0A0A1XI69 A0A0A1WUY1 A0A1I8M5A8 A0A0K8UL76 B3MNS5 B4N0Y9 A0A0K8WAJ4 E2AIM4 A0A212EPM6 A0A232F8C9 A0A026WVQ0 A0A3L8DFK2 A0A1J1IQ95 B4JAK8 A0A2P8YQG3 D6WBV6 A0A158NQH7 K7IR20 A0A151X5R7 E9IDK7 A0A1W4X7R5 A0A0A1XE33 A0A1J1IPR1 A0A195CQD8 A0A1J0M5H9 J9JWA0
Pubmed
28756777
26354079
20966253
12364791
14747013
17210077
+ More
25244985 20920257 24945155 26483478 19121390 17510324 22651552 17994087 15632085 23185243 20075255 28648823 24495485 25348373 24330624 18057021 25830018 25315136 20798317 22118469 24508170 30249741 29403074 18362917 19820115 21347285 21282665
25244985 20920257 24945155 26483478 19121390 17510324 22651552 17994087 15632085 23185243 20075255 28648823 24495485 25348373 24330624 18057021 25830018 25315136 20798317 22118469 24508170 30249741 29403074 18362917 19820115 21347285 21282665
EMBL
NWSH01003717
PCG65891.1
KZ150304
PZC71518.1
ODYU01007212
SOQ49795.1
+ More
KQ459589 KPI97640.1 KQ460367 KPJ15487.1 RSAL01000070 RVE49162.1 DS231860 EDS39335.1 AAAB01008984 EAL39045.3 APCN01003737 AXCM01002168 GANO01004167 JAB55704.1 UFQT01001144 SSX29131.1 GFDF01007636 JAV06448.1 AXCN02000839 AJWK01030620 ODYU01001075 SOQ36804.1 ODYU01000288 SOQ34781.1 GGFJ01007734 MBW56875.1 GGFM01003569 MBW24320.1 GAPW01002677 JAC10921.1 GAPW01002676 JAC10922.1 KQ460883 KPJ11225.1 KQ459603 KPI92731.1 GGFJ01007342 MBW56483.1 JXUM01071083 KQ562644 KXJ75422.1 GGFK01008407 MBW41728.1 GGFK01008113 MBW41434.1 GGFK01008017 MBW41338.1 GGFL01000581 MBW64759.1 GAMD01000738 JAB00853.1 BABH01005602 UFQS01000438 UFQT01000438 SSX03949.1 SSX24314.1 GGFL01003854 MBW68032.1 CH477415 EAT41378.1 EJY57664.1 EAT41380.1 EAT41382.1 SSX29130.1 AK403208 BAM19633.1 CVRI01000063 CRL04727.1 CH479180 EDW28518.1 CH379060 EAL33455.2 KRT03851.1 AJVK01011714 AJVK01011715 OUUW01000010 SPP86199.1 AAZX01007716 NNAY01001349 OXU24282.1 GAMC01010223 JAB96332.1 GAKP01015713 JAC43239.1 GALA01000092 JAA94760.1 EJY57663.1 KQ459580 KPI99251.1 CH933807 EDW11824.1 KRG02930.1 KRG02931.1 KRG02932.1 KQ460240 KPJ16392.1 CH940649 EDW63706.1 KA648010 AFP62639.1 GBXI01003635 JAD10657.1 GBXI01012054 JAD02238.1 GDHF01024860 GDHF01014589 GDHF01004371 JAI27454.1 JAI37725.1 JAI47943.1 CH902620 EDV31162.1 KPU73246.1 CH963920 EDW78151.1 GDHF01004464 JAI47850.1 GL439840 EFN66713.1 AGBW02013442 OWR43429.1 NNAY01000669 OXU27086.1 KK107109 EZA59174.1 QOIP01000008 RLU19237.1 CVRI01000056 CRL01708.1 CH916368 EDW02794.1 PYGN01000432 PSN46485.1 KQ971315 EEZ97864.2 ADTU01023297 ADTU01023298 ADTU01023299 ADTU01023300 ADTU01023301 ADTU01023302 ADTU01023303 ADTU01023304 KQ982494 KYQ55731.1 GL762454 EFZ21361.1 GBXI01007275 GBXI01005070 GBXI01004777 JAD07017.1 JAD09222.1 JAD09515.1 CRL01706.1 KQ977444 KYN02692.1 KU663628 APD15622.1 ABLF02035114 ABLF02035118
KQ459589 KPI97640.1 KQ460367 KPJ15487.1 RSAL01000070 RVE49162.1 DS231860 EDS39335.1 AAAB01008984 EAL39045.3 APCN01003737 AXCM01002168 GANO01004167 JAB55704.1 UFQT01001144 SSX29131.1 GFDF01007636 JAV06448.1 AXCN02000839 AJWK01030620 ODYU01001075 SOQ36804.1 ODYU01000288 SOQ34781.1 GGFJ01007734 MBW56875.1 GGFM01003569 MBW24320.1 GAPW01002677 JAC10921.1 GAPW01002676 JAC10922.1 KQ460883 KPJ11225.1 KQ459603 KPI92731.1 GGFJ01007342 MBW56483.1 JXUM01071083 KQ562644 KXJ75422.1 GGFK01008407 MBW41728.1 GGFK01008113 MBW41434.1 GGFK01008017 MBW41338.1 GGFL01000581 MBW64759.1 GAMD01000738 JAB00853.1 BABH01005602 UFQS01000438 UFQT01000438 SSX03949.1 SSX24314.1 GGFL01003854 MBW68032.1 CH477415 EAT41378.1 EJY57664.1 EAT41380.1 EAT41382.1 SSX29130.1 AK403208 BAM19633.1 CVRI01000063 CRL04727.1 CH479180 EDW28518.1 CH379060 EAL33455.2 KRT03851.1 AJVK01011714 AJVK01011715 OUUW01000010 SPP86199.1 AAZX01007716 NNAY01001349 OXU24282.1 GAMC01010223 JAB96332.1 GAKP01015713 JAC43239.1 GALA01000092 JAA94760.1 EJY57663.1 KQ459580 KPI99251.1 CH933807 EDW11824.1 KRG02930.1 KRG02931.1 KRG02932.1 KQ460240 KPJ16392.1 CH940649 EDW63706.1 KA648010 AFP62639.1 GBXI01003635 JAD10657.1 GBXI01012054 JAD02238.1 GDHF01024860 GDHF01014589 GDHF01004371 JAI27454.1 JAI37725.1 JAI47943.1 CH902620 EDV31162.1 KPU73246.1 CH963920 EDW78151.1 GDHF01004464 JAI47850.1 GL439840 EFN66713.1 AGBW02013442 OWR43429.1 NNAY01000669 OXU27086.1 KK107109 EZA59174.1 QOIP01000008 RLU19237.1 CVRI01000056 CRL01708.1 CH916368 EDW02794.1 PYGN01000432 PSN46485.1 KQ971315 EEZ97864.2 ADTU01023297 ADTU01023298 ADTU01023299 ADTU01023300 ADTU01023301 ADTU01023302 ADTU01023303 ADTU01023304 KQ982494 KYQ55731.1 GL762454 EFZ21361.1 GBXI01007275 GBXI01005070 GBXI01004777 JAD07017.1 JAD09222.1 JAD09515.1 CRL01706.1 KQ977444 KYN02692.1 KU663628 APD15622.1 ABLF02035114 ABLF02035118
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000002320
UP000075882
+ More
UP000007062 UP000075840 UP000075903 UP000075902 UP000076407 UP000075901 UP000075920 UP000076408 UP000075883 UP000075886 UP000075900 UP000092461 UP000075885 UP000069272 UP000069940 UP000249989 UP000075880 UP000005204 UP000008820 UP000183832 UP000008744 UP000001819 UP000092462 UP000268350 UP000075884 UP000002358 UP000215335 UP000009192 UP000095300 UP000008792 UP000095301 UP000007801 UP000007798 UP000000311 UP000007151 UP000053097 UP000279307 UP000001070 UP000245037 UP000007266 UP000005205 UP000075809 UP000192223 UP000078542 UP000007819
UP000007062 UP000075840 UP000075903 UP000075902 UP000076407 UP000075901 UP000075920 UP000076408 UP000075883 UP000075886 UP000075900 UP000092461 UP000075885 UP000069272 UP000069940 UP000249989 UP000075880 UP000005204 UP000008820 UP000183832 UP000008744 UP000001819 UP000092462 UP000268350 UP000075884 UP000002358 UP000215335 UP000009192 UP000095300 UP000008792 UP000095301 UP000007801 UP000007798 UP000000311 UP000007151 UP000053097 UP000279307 UP000001070 UP000245037 UP000007266 UP000005205 UP000075809 UP000192223 UP000078542 UP000007819
PRIDE
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2A4J1M9
A0A2W1B7Q2
A0A2H1W9L5
A0A194Q2K6
A0A194RCZ1
A0A3S2NUP7
+ More
B0W8V6 A0A182KUQ0 Q5TNF5 A0A182I4Y9 A0A1S4H040 A0A182VEG6 A0A1S4H0X8 A0A182TXT4 A0A182X2W4 A0A182SXZ4 A0A182WGZ2 A0A182YFE7 A0A182MGV1 U5EQP3 A0A336MI67 A0A1L8DJ03 A0A182QEE4 A0A182R9P9 A0A1B0CVI8 A0A2H1V975 A0A182PDD3 A0A2H1V1S5 A0A2M4BUY8 A0A2M3Z754 A0A2K6VB47 A0A182F5H0 A0A023EPI5 A0A023END9 A0A194R1U8 A0A194PH51 A0A2M4BU04 A0A182GL29 A0A182IUI2 A0A2M4AM81 A0A2M4AKZ7 A0A2M4AL49 A0A2M4CHM7 T1EAU4 H9JLK0 A0A336KHN4 A0A2M4CRR7 Q173Z1 Q173Z2 A0A336MGQ6 I4DNZ3 A0A1J1IX47 B4G838 Q29N87 A0A1B0D5A4 A0A3B0KIF6 A0A182NEW7 K7J451 A0A232F124 W8BCB3 A0A034VLB9 T1DGR4 J9HSR1 A0A194Q121 B4KE79 A0A1I8PDV7 A0A0N1PJG6 B4LS98 T1PIS7 A0A0A1XI69 A0A0A1WUY1 A0A1I8M5A8 A0A0K8UL76 B3MNS5 B4N0Y9 A0A0K8WAJ4 E2AIM4 A0A212EPM6 A0A232F8C9 A0A026WVQ0 A0A3L8DFK2 A0A1J1IQ95 B4JAK8 A0A2P8YQG3 D6WBV6 A0A158NQH7 K7IR20 A0A151X5R7 E9IDK7 A0A1W4X7R5 A0A0A1XE33 A0A1J1IPR1 A0A195CQD8 A0A1J0M5H9 J9JWA0
B0W8V6 A0A182KUQ0 Q5TNF5 A0A182I4Y9 A0A1S4H040 A0A182VEG6 A0A1S4H0X8 A0A182TXT4 A0A182X2W4 A0A182SXZ4 A0A182WGZ2 A0A182YFE7 A0A182MGV1 U5EQP3 A0A336MI67 A0A1L8DJ03 A0A182QEE4 A0A182R9P9 A0A1B0CVI8 A0A2H1V975 A0A182PDD3 A0A2H1V1S5 A0A2M4BUY8 A0A2M3Z754 A0A2K6VB47 A0A182F5H0 A0A023EPI5 A0A023END9 A0A194R1U8 A0A194PH51 A0A2M4BU04 A0A182GL29 A0A182IUI2 A0A2M4AM81 A0A2M4AKZ7 A0A2M4AL49 A0A2M4CHM7 T1EAU4 H9JLK0 A0A336KHN4 A0A2M4CRR7 Q173Z1 Q173Z2 A0A336MGQ6 I4DNZ3 A0A1J1IX47 B4G838 Q29N87 A0A1B0D5A4 A0A3B0KIF6 A0A182NEW7 K7J451 A0A232F124 W8BCB3 A0A034VLB9 T1DGR4 J9HSR1 A0A194Q121 B4KE79 A0A1I8PDV7 A0A0N1PJG6 B4LS98 T1PIS7 A0A0A1XI69 A0A0A1WUY1 A0A1I8M5A8 A0A0K8UL76 B3MNS5 B4N0Y9 A0A0K8WAJ4 E2AIM4 A0A212EPM6 A0A232F8C9 A0A026WVQ0 A0A3L8DFK2 A0A1J1IQ95 B4JAK8 A0A2P8YQG3 D6WBV6 A0A158NQH7 K7IR20 A0A151X5R7 E9IDK7 A0A1W4X7R5 A0A0A1XE33 A0A1J1IPR1 A0A195CQD8 A0A1J0M5H9 J9JWA0
PDB
1N8S
E-value=4.96308e-16,
Score=205
Ontologies
GO
PANTHER
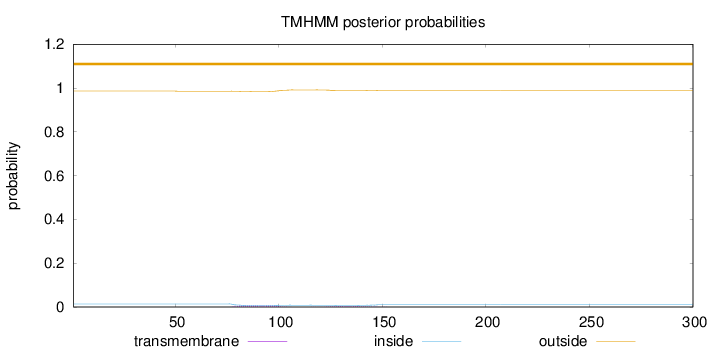
Topology
Subcellular location
Secreted
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25683
Exp number, first 60 AAs:
0.00065
Total prob of N-in:
0.01407
outside
1 - 300
Population Genetic Test Statistics
Pi
232.96658
Theta
158.738862
Tajima's D
1.382109
CLR
0.000002
CSRT
0.758562071896405
Interpretation
Uncertain
