Pre Gene Modal
BGIBMGA008730
Annotation
PREDICTED:_synapse-associated_protein_of_47_kDa_[Bombyx_mori]
Full name
Synapse-associated protein of 47 kDa
Location in the cell
Cytoplasmic Reliability : 0.794
Sequence
CDS
ATGCACATGCTCTGTGTAAAGTTTGCCATCCAAGCTCTATCGCAAATGAGCTTCCATTCCGATATAATGAGCATCTTGGGTGAATTCAATCGCGAGCAGGACGCCTTCATCAAAGGACAAGAGAAAGCCGGTGCTGGGTCGGCTATTGCTCCCTGGATCGGCGCTCCTAACGAAGCTGCTCTGAAGGAAGAATGCCTCTCCCTTTCCACTGATCGAAGGAACTTTGTCCGCGCTCCGCCGGCGGGCGTGGAGTTCGACTTCGATTACGACAAGATGTACCCGGTGGCGGTCGCAATTATGACCGAGGACCCGAACCTTGAGAAGATGCGCTTTGATCTTGTGCCAAAAGTTATCACTGAAGAGAACTTTTGGAGGAACTACTTCTACCGCCTGTCGCTGATCTGCCAGGCCAACGAGGCCGACGCCCTCGCGGCGCGCCATTCCTCCGCCGACGACTCCCAGGCTTAA
Protein
MHMLCVKFAIQALSQMSFHSDIMSILGEFNREQDAFIKGQEKAGAGSAIAPWIGAPNEAALKEECLSLSTDRRNFVRAPPAGVEFDFDYDKMYPVAVAIMTEDPNLEKMRFDLVPKVITEENFWRNYFYRLSLICQANEADALAARHSSADDSQA
Summary
Keywords
Alternative splicing
Complete proteome
Phosphoprotein
Reference proteome
Feature
chain Synapse-associated protein of 47 kDa
splice variant In isoform F, isoform G and isoform I.
splice variant In isoform F, isoform G and isoform I.
Uniprot
H9JGT3
A0A2H1VP24
A0A194PW87
A0A194RCB8
A0A3S2LSW5
A0A212F533
+ More
A0A1E1W4Q7 S4PW91 A0A2W1B5S7 A0A2A4K4F3 A0A2A4K3V9 A0A2A4K4W3 A0A1W4XB29 A0A0A9W017 A0A0A9XNQ4 A0A0A9XMR6 A0A0A9XKK8 A0A0K8S9K3 A0A146LH73 A0A0A9XQZ0 A0A0A9XMS2 A0A0T6B916 A0A146LFW7 A0A1B6G6M2 D6W7Y3 A0A182PM84 A0A1B6C4T4 A0A1Y1K6Y0 A0A1Y1K415 A0A139WPJ7 A0A182W167 A0A182SDQ0 A0A182XG09 A0A1B6CA49 V5GV31 A0A1Y1K6U5 A0A1Y1K7P2 A0A1B6CPZ5 A0A1B6JJR0 A0A1B6HPM1 A0A1B6D2G3 A0A067QS14 A0A182FPG0 A0A1B6K2S3 A0A1B6K805 A0A1B6CJQ8 A0A182ID55 A0A182KCI5 A0A2J7PGZ2 A0A2R7VXW3 Q173T4 A0A182UQ59 F5HL82 E0VK05 N6T9F4 A0A182L6Z9 A0A1B6G4E8 A0A1B6FPQ6 A0A182H6G4 A0A182GTY3 Q173T6 A0A182XX30 A0A182JCR9 A0A084WP12 A0A1Q3FCF7 A0A2M4CIA0 A0A2M4CI67 Q173T5 A0A023EV36 A0A2M4BKY2 A0A2M4BKT4 A0A1B6LES7 A0A2M4AQQ6 A0A1B6LWB3 F5HL80 A0A069DS44 A0A0P4VTJ0 A0A2M4AMF0 A0A1B6KTM1 A0A1B6KKR3 A0A224XUK1 A0A182MKH8 A0A2S2QHD5 A0A1B6MHT8 A0A1B6KPY6 A0A0C9PU23 A0A0R1E7U0 Q960T2-4 A0A1B0BBV9 W5JGS8 A0A0K8WHY5 A0A1B6E6R5 T1P7T5 F3YDF6 A0A0R1E777 Q960T2-3 A0A0B4LH71 A0A1A9ZTC1 Q960T2-2 T1PK00
A0A1E1W4Q7 S4PW91 A0A2W1B5S7 A0A2A4K4F3 A0A2A4K3V9 A0A2A4K4W3 A0A1W4XB29 A0A0A9W017 A0A0A9XNQ4 A0A0A9XMR6 A0A0A9XKK8 A0A0K8S9K3 A0A146LH73 A0A0A9XQZ0 A0A0A9XMS2 A0A0T6B916 A0A146LFW7 A0A1B6G6M2 D6W7Y3 A0A182PM84 A0A1B6C4T4 A0A1Y1K6Y0 A0A1Y1K415 A0A139WPJ7 A0A182W167 A0A182SDQ0 A0A182XG09 A0A1B6CA49 V5GV31 A0A1Y1K6U5 A0A1Y1K7P2 A0A1B6CPZ5 A0A1B6JJR0 A0A1B6HPM1 A0A1B6D2G3 A0A067QS14 A0A182FPG0 A0A1B6K2S3 A0A1B6K805 A0A1B6CJQ8 A0A182ID55 A0A182KCI5 A0A2J7PGZ2 A0A2R7VXW3 Q173T4 A0A182UQ59 F5HL82 E0VK05 N6T9F4 A0A182L6Z9 A0A1B6G4E8 A0A1B6FPQ6 A0A182H6G4 A0A182GTY3 Q173T6 A0A182XX30 A0A182JCR9 A0A084WP12 A0A1Q3FCF7 A0A2M4CIA0 A0A2M4CI67 Q173T5 A0A023EV36 A0A2M4BKY2 A0A2M4BKT4 A0A1B6LES7 A0A2M4AQQ6 A0A1B6LWB3 F5HL80 A0A069DS44 A0A0P4VTJ0 A0A2M4AMF0 A0A1B6KTM1 A0A1B6KKR3 A0A224XUK1 A0A182MKH8 A0A2S2QHD5 A0A1B6MHT8 A0A1B6KPY6 A0A0C9PU23 A0A0R1E7U0 Q960T2-4 A0A1B0BBV9 W5JGS8 A0A0K8WHY5 A0A1B6E6R5 T1P7T5 F3YDF6 A0A0R1E777 Q960T2-3 A0A0B4LH71 A0A1A9ZTC1 Q960T2-2 T1PK00
Pubmed
19121390
26354079
22118469
23622113
28756777
25401762
+ More
26823975 18362917 19820115 28004739 24845553 17510324 12364791 14747013 17210077 20566863 23537049 20966253 26483478 25244985 24438588 24945155 26334808 27129103 17994087 17550304 7494462 10731132 12537572 12537569 18327897 20920257 23761445 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356
26823975 18362917 19820115 28004739 24845553 17510324 12364791 14747013 17210077 20566863 23537049 20966253 26483478 25244985 24438588 24945155 26334808 27129103 17994087 17550304 7494462 10731132 12537572 12537569 18327897 20920257 23761445 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01034414
BABH01034415
ODYU01003353
SOQ42004.1
KQ459589
KPI97641.1
+ More
KQ460367 KPJ15488.1 RSAL01000271 RVE43165.1 AGBW02010265 OWR48851.1 GDQN01009193 JAT81861.1 GAIX01007198 JAA85362.1 KZ150304 PZC71519.1 NWSH01000158 PCG78949.1 PCG78947.1 PCG78948.1 GBHO01043811 GBRD01016398 JAF99792.1 JAG49428.1 GBHO01022338 JAG21266.1 GBHO01022345 JAG21259.1 GBHO01022342 GBRD01016395 JAG21262.1 JAG49431.1 GBRD01016396 JAG49430.1 GDHC01011750 JAQ06879.1 GBHO01022341 JAG21263.1 GBHO01022335 GBRD01015389 JAG21269.1 JAG50437.1 LJIG01009302 KRT83351.1 GDHC01011476 JAQ07153.1 GECZ01011681 JAS58088.1 KQ971307 EFA11027.1 GEDC01028777 JAS08521.1 GEZM01093619 JAV56218.1 GEZM01093617 JAV56219.1 KYB29817.1 GEDC01027168 JAS10130.1 GALX01002939 JAB65527.1 GEZM01093615 JAV56221.1 GEZM01093616 JAV56220.1 GEDC01021739 JAS15559.1 GECU01008291 JAS99415.1 GECU01031161 JAS76545.1 GEDC01017495 JAS19803.1 KK853022 KDR12364.1 GECU01013924 GECU01001934 JAS93782.1 JAT05773.1 GECU01023619 GECU01000393 JAS84087.1 JAT07314.1 GEDC01023733 JAS13565.1 APCN01005030 NEVH01025158 PNF15600.1 KK854149 PTY12179.1 CH477416 EAT41364.1 AAAB01008880 EGK97041.1 DS235235 EEB13711.1 APGK01039172 KB740967 KB632399 ENN76884.1 ERL94628.1 GECZ01026628 GECZ01015277 GECZ01013618 GECZ01012463 GECZ01006200 JAS43141.1 JAS54492.1 JAS56151.1 JAS57306.1 JAS63569.1 GECZ01017574 JAS52195.1 JXUM01027088 JXUM01027089 KQ560777 KXJ80933.1 JXUM01088159 JXUM01088160 JXUM01088161 JXUM01088162 JXUM01088163 KQ563684 KXJ73495.1 EAT41363.1 ATLV01024813 ATLV01024814 ATLV01024815 KE525361 KFB51956.1 GFDL01009781 JAV25264.1 GGFL01000855 MBW65033.1 GGFL01000856 MBW65034.1 EAT41365.1 GAPW01001254 JAC12344.1 GGFJ01004589 MBW53730.1 GGFJ01004539 MBW53680.1 GEBQ01017817 JAT22160.1 GGFK01009793 MBW43114.1 GEBQ01011995 JAT27982.1 EGK97043.1 GBGD01002036 JAC86853.1 GDKW01000018 JAI56577.1 GGFK01008630 MBW41951.1 GEBQ01025209 JAT14768.1 GEBQ01027944 JAT12033.1 GFTR01004326 JAW12100.1 AXCM01000384 GGMS01007962 MBY77165.1 GEBQ01004447 JAT35530.1 GEBQ01026490 JAT13487.1 GBYB01004833 JAG74600.1 CM000160 KRK03714.1 X80110 X80111 AE014297 AY051871 JXJN01011702 JXJN01011703 JXJN01011704 JXJN01011705 ADMH02001267 ETN63291.1 GDHF01003214 GDHF01001829 JAI49100.1 JAI50485.1 GEDC01003678 JAS33620.1 KA644597 AFP59226.1 BT126317 AEB41030.1 AGB95997.1 KRK03713.1 AHN57349.1 KA649091 AFP63720.1
KQ460367 KPJ15488.1 RSAL01000271 RVE43165.1 AGBW02010265 OWR48851.1 GDQN01009193 JAT81861.1 GAIX01007198 JAA85362.1 KZ150304 PZC71519.1 NWSH01000158 PCG78949.1 PCG78947.1 PCG78948.1 GBHO01043811 GBRD01016398 JAF99792.1 JAG49428.1 GBHO01022338 JAG21266.1 GBHO01022345 JAG21259.1 GBHO01022342 GBRD01016395 JAG21262.1 JAG49431.1 GBRD01016396 JAG49430.1 GDHC01011750 JAQ06879.1 GBHO01022341 JAG21263.1 GBHO01022335 GBRD01015389 JAG21269.1 JAG50437.1 LJIG01009302 KRT83351.1 GDHC01011476 JAQ07153.1 GECZ01011681 JAS58088.1 KQ971307 EFA11027.1 GEDC01028777 JAS08521.1 GEZM01093619 JAV56218.1 GEZM01093617 JAV56219.1 KYB29817.1 GEDC01027168 JAS10130.1 GALX01002939 JAB65527.1 GEZM01093615 JAV56221.1 GEZM01093616 JAV56220.1 GEDC01021739 JAS15559.1 GECU01008291 JAS99415.1 GECU01031161 JAS76545.1 GEDC01017495 JAS19803.1 KK853022 KDR12364.1 GECU01013924 GECU01001934 JAS93782.1 JAT05773.1 GECU01023619 GECU01000393 JAS84087.1 JAT07314.1 GEDC01023733 JAS13565.1 APCN01005030 NEVH01025158 PNF15600.1 KK854149 PTY12179.1 CH477416 EAT41364.1 AAAB01008880 EGK97041.1 DS235235 EEB13711.1 APGK01039172 KB740967 KB632399 ENN76884.1 ERL94628.1 GECZ01026628 GECZ01015277 GECZ01013618 GECZ01012463 GECZ01006200 JAS43141.1 JAS54492.1 JAS56151.1 JAS57306.1 JAS63569.1 GECZ01017574 JAS52195.1 JXUM01027088 JXUM01027089 KQ560777 KXJ80933.1 JXUM01088159 JXUM01088160 JXUM01088161 JXUM01088162 JXUM01088163 KQ563684 KXJ73495.1 EAT41363.1 ATLV01024813 ATLV01024814 ATLV01024815 KE525361 KFB51956.1 GFDL01009781 JAV25264.1 GGFL01000855 MBW65033.1 GGFL01000856 MBW65034.1 EAT41365.1 GAPW01001254 JAC12344.1 GGFJ01004589 MBW53730.1 GGFJ01004539 MBW53680.1 GEBQ01017817 JAT22160.1 GGFK01009793 MBW43114.1 GEBQ01011995 JAT27982.1 EGK97043.1 GBGD01002036 JAC86853.1 GDKW01000018 JAI56577.1 GGFK01008630 MBW41951.1 GEBQ01025209 JAT14768.1 GEBQ01027944 JAT12033.1 GFTR01004326 JAW12100.1 AXCM01000384 GGMS01007962 MBY77165.1 GEBQ01004447 JAT35530.1 GEBQ01026490 JAT13487.1 GBYB01004833 JAG74600.1 CM000160 KRK03714.1 X80110 X80111 AE014297 AY051871 JXJN01011702 JXJN01011703 JXJN01011704 JXJN01011705 ADMH02001267 ETN63291.1 GDHF01003214 GDHF01001829 JAI49100.1 JAI50485.1 GEDC01003678 JAS33620.1 KA644597 AFP59226.1 BT126317 AEB41030.1 AGB95997.1 KRK03713.1 AHN57349.1 KA649091 AFP63720.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000218220
+ More
UP000192223 UP000007266 UP000075885 UP000075920 UP000075901 UP000076407 UP000027135 UP000069272 UP000075840 UP000075881 UP000235965 UP000008820 UP000075903 UP000007062 UP000009046 UP000019118 UP000030742 UP000075882 UP000069940 UP000249989 UP000076408 UP000075880 UP000030765 UP000075883 UP000002282 UP000000803 UP000092460 UP000000673 UP000092445
UP000192223 UP000007266 UP000075885 UP000075920 UP000075901 UP000076407 UP000027135 UP000069272 UP000075840 UP000075881 UP000235965 UP000008820 UP000075903 UP000007062 UP000009046 UP000019118 UP000030742 UP000075882 UP000069940 UP000249989 UP000076408 UP000075880 UP000030765 UP000075883 UP000002282 UP000000803 UP000092460 UP000000673 UP000092445
Pfam
PF03909 BSD
Gene 3D
ProteinModelPortal
H9JGT3
A0A2H1VP24
A0A194PW87
A0A194RCB8
A0A3S2LSW5
A0A212F533
+ More
A0A1E1W4Q7 S4PW91 A0A2W1B5S7 A0A2A4K4F3 A0A2A4K3V9 A0A2A4K4W3 A0A1W4XB29 A0A0A9W017 A0A0A9XNQ4 A0A0A9XMR6 A0A0A9XKK8 A0A0K8S9K3 A0A146LH73 A0A0A9XQZ0 A0A0A9XMS2 A0A0T6B916 A0A146LFW7 A0A1B6G6M2 D6W7Y3 A0A182PM84 A0A1B6C4T4 A0A1Y1K6Y0 A0A1Y1K415 A0A139WPJ7 A0A182W167 A0A182SDQ0 A0A182XG09 A0A1B6CA49 V5GV31 A0A1Y1K6U5 A0A1Y1K7P2 A0A1B6CPZ5 A0A1B6JJR0 A0A1B6HPM1 A0A1B6D2G3 A0A067QS14 A0A182FPG0 A0A1B6K2S3 A0A1B6K805 A0A1B6CJQ8 A0A182ID55 A0A182KCI5 A0A2J7PGZ2 A0A2R7VXW3 Q173T4 A0A182UQ59 F5HL82 E0VK05 N6T9F4 A0A182L6Z9 A0A1B6G4E8 A0A1B6FPQ6 A0A182H6G4 A0A182GTY3 Q173T6 A0A182XX30 A0A182JCR9 A0A084WP12 A0A1Q3FCF7 A0A2M4CIA0 A0A2M4CI67 Q173T5 A0A023EV36 A0A2M4BKY2 A0A2M4BKT4 A0A1B6LES7 A0A2M4AQQ6 A0A1B6LWB3 F5HL80 A0A069DS44 A0A0P4VTJ0 A0A2M4AMF0 A0A1B6KTM1 A0A1B6KKR3 A0A224XUK1 A0A182MKH8 A0A2S2QHD5 A0A1B6MHT8 A0A1B6KPY6 A0A0C9PU23 A0A0R1E7U0 Q960T2-4 A0A1B0BBV9 W5JGS8 A0A0K8WHY5 A0A1B6E6R5 T1P7T5 F3YDF6 A0A0R1E777 Q960T2-3 A0A0B4LH71 A0A1A9ZTC1 Q960T2-2 T1PK00
A0A1E1W4Q7 S4PW91 A0A2W1B5S7 A0A2A4K4F3 A0A2A4K3V9 A0A2A4K4W3 A0A1W4XB29 A0A0A9W017 A0A0A9XNQ4 A0A0A9XMR6 A0A0A9XKK8 A0A0K8S9K3 A0A146LH73 A0A0A9XQZ0 A0A0A9XMS2 A0A0T6B916 A0A146LFW7 A0A1B6G6M2 D6W7Y3 A0A182PM84 A0A1B6C4T4 A0A1Y1K6Y0 A0A1Y1K415 A0A139WPJ7 A0A182W167 A0A182SDQ0 A0A182XG09 A0A1B6CA49 V5GV31 A0A1Y1K6U5 A0A1Y1K7P2 A0A1B6CPZ5 A0A1B6JJR0 A0A1B6HPM1 A0A1B6D2G3 A0A067QS14 A0A182FPG0 A0A1B6K2S3 A0A1B6K805 A0A1B6CJQ8 A0A182ID55 A0A182KCI5 A0A2J7PGZ2 A0A2R7VXW3 Q173T4 A0A182UQ59 F5HL82 E0VK05 N6T9F4 A0A182L6Z9 A0A1B6G4E8 A0A1B6FPQ6 A0A182H6G4 A0A182GTY3 Q173T6 A0A182XX30 A0A182JCR9 A0A084WP12 A0A1Q3FCF7 A0A2M4CIA0 A0A2M4CI67 Q173T5 A0A023EV36 A0A2M4BKY2 A0A2M4BKT4 A0A1B6LES7 A0A2M4AQQ6 A0A1B6LWB3 F5HL80 A0A069DS44 A0A0P4VTJ0 A0A2M4AMF0 A0A1B6KTM1 A0A1B6KKR3 A0A224XUK1 A0A182MKH8 A0A2S2QHD5 A0A1B6MHT8 A0A1B6KPY6 A0A0C9PU23 A0A0R1E7U0 Q960T2-4 A0A1B0BBV9 W5JGS8 A0A0K8WHY5 A0A1B6E6R5 T1P7T5 F3YDF6 A0A0R1E777 Q960T2-3 A0A0B4LH71 A0A1A9ZTC1 Q960T2-2 T1PK00
PDB
1X3A
E-value=1.19472e-25,
Score=283
Ontologies
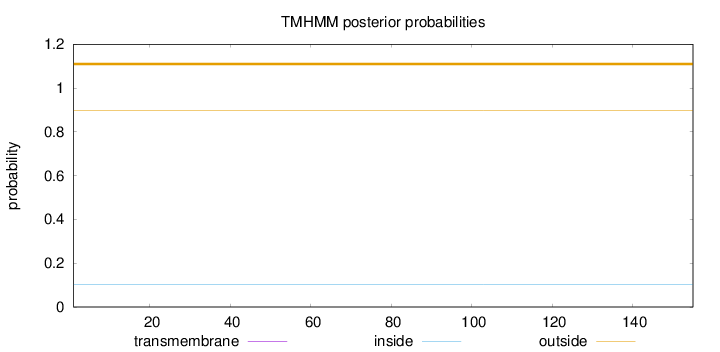
Topology
Length:
155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00248
Exp number, first 60 AAs:
0.00188
Total prob of N-in:
0.10158
outside
1 - 155
Population Genetic Test Statistics
Pi
158.675381
Theta
156.082966
Tajima's D
-0.558631
CLR
22.247119
CSRT
0.227388630568472
Interpretation
Uncertain
