Gene
KWMTBOMO04186
Pre Gene Modal
BGIBMGA000182
Annotation
PREDICTED:_organic_cation_transporter_protein-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.726
Sequence
CDS
ATGGACGGTACGAAACGGAAATCGGTAACTATTTCTGAAAAAATAGAAGATACACCGAAACAAGATGAAGATAAACTGATTCAAGCGATAGGTGAATTCGGGAGATGGCACGTGATGCTAACTATGACGCTATCTGCACCGATTCGAATGACGGCAGTCTGGAACCATTTGGGAATCATATTTCTGGCACCGAAAACTGAATTTATTTGCGCGGAGAGACGTAATAACAACGAAACCATCCAAGAATCGGAGTGCTACAGTGATTGTACTGAATACGAATACAAAACGGATTTCGTGAATACAATCATTTCTGAATGGGACCTAGTTTGCGATCGATCCTGGCTGACAAACTTCAACCAGACAGCTTGCATGTCCGGTGTTTTATTGGGCAGTATAGTTTTTGGATTTTTCGCTGATAGATTCGGTCGTCGTCCCGCTCTCCTGACGGCCTGCACCACGCAACTGATAGCTGGATTCACAGCGCCATTTTGCCCAAATTACATCACATTCACCTTGGTCCGATTCTTCAACGGCGCAGCTGTTGCTGGCGCATTACTGACTTCTTTTATTCTAATGATTGAAACCACGGGCATATCGAAAAGGGAACTAATGAACTGTATCGTTTCCGTGCCTCTGTCGGTGGGAGAAATGATAATGCCAATGTTTTCATATTACTTGAGGTCTTGGACTAGATATAGTTTTGGTCTCGCCCTGCCAAATTTGGTATTTTTGGTTTTATTCTTCATCGTACCAGAGTCTCCGAAGTGGTTGATTTCCGTTGGAAGGCTCGAGGAGGCGAGTTTGATTATGACGAAAATAGCGAAATGGAATAGACTTCCAACAGAAGACATGATGGATAGAGTAAAGATTATAGCATTTGAATCCAATGTTGACGCCCACAAAAACGTCAAGGCTACTTACTTAGACCTGATCAAAGTGCAGGAGATAAGGTGGAAGAATATCGGATGCTGTCTTACGTGGTTTATCCTCGGGCTGAGCTTCTATGGCAGCAATCAGTACATAGGACAGACCAGCCCTAATGTTTTCGTTGCTATTGCAATGGCTGGAGCTATTCAGACTACCTTTGACCCATTCCTTGTTCTATTACTGACCATAGAGACCGACTTAGCTGGAGGTCTTGAGGCTAATGAACTGATCATAATCGGCCACACGTTAACAGCCAAAAAGTAG
Protein
MDGTKRKSVTISEKIEDTPKQDEDKLIQAIGEFGRWHVMLTMTLSAPIRMTAVWNHLGIIFLAPKTEFICAERRNNNETIQESECYSDCTEYEYKTDFVNTIISEWDLVCDRSWLTNFNQTACMSGVLLGSIVFGFFADRFGRRPALLTACTTQLIAGFTAPFCPNYITFTLVRFFNGAAVAGALLTSFILMIETTGISKRELMNCIVSVPLSVGEMIMPMFSYYLRSWTRYSFGLALPNLVFLVLFFIVPESPKWLISVGRLEEASLIMTKIAKWNRLPTEDMMDRVKIIAFESNVDAHKNVKATYLDLIKVQEIRWKNIGCCLTWFILGLSFYGSNQYIGQTSPNVFVAIAMAGAIQTTFDPFLVLLLTIETDLAGGLEANELIIIGHTLTAKK
Summary
Uniprot
H9ISF5
H9JGL2
A0A2H1VMD7
A0A2W1BFH6
A0A0L7LUI9
A0A2A4K5L0
+ More
A0A194RHR5 A0A2A4J3B3 A0A2W1BP27 A0A212F8L3 A0A2H1VGH7 I4DNF7 A0A194PMC3 H9JNB3 A0A2A4KA95 A0A194QN55 A0A2A4K9Y1 A0A194PY14 A0A1J1IHN1 A0A1B0C9J9 A0A2A4K9Y5 A0A2H1W3H1 A0A1Q3FH65 V5IAH1 D6WKJ1 B0X4Q4 A0A1Y1KD57 A0A1Y1K9J5 Q17CS1 D6WKJ0 A0A088ALA3 A0A023EUG3 A0A182H4N7 A0A0L7QRN1 A0A182T1C6 H9JNI6 A0A154P8V9 A0A182PAB8 A0A182Y4D6 A0A2W1BCH7 A0A182VI17 A0A2A3ESR9 K7ITG9 A0A182TEQ3 A0A182QLT3 A0A2H1VUP3 A0A0M9A4G4 A0A182WRH0 Q7QDB3 A0A182HUA7 D6X4T8 A0A2M4CTR9 A0A2M4CU13 A0A182W090 A0A182K794 A0A310SN29 B3NK59 A0A2J7RBY0 B4HPT1 A0A194QK42 U4UII8 A0A0M9A1C2 N6T164 A0A182MBN7 A0A182JCT1 A0A182NFM5 Q7K3M6 B4PAQ3 B4QEI8 E2BIH0 A0A232F564 A0A182RNN4 A0A2A4J566 A0A182F7G4 A0A212FFR4 A0A2W1BI79 A0A195DRF1 A0A194QIQ7 B3MH68 A0A088A3V4 B4MRV4 A0A0M3QVP8 K7JAK3 A0A1W4VGV5 A0A084VXP9 A0A310SUT5 E2A1J5 A0A1B6EKM5 A0A026W714 A0A158NPS8 A0A195ATZ5 B4JWE6 A0A0L7LUK1 A0A195FCZ0 A0A232EL12 E1ZZY2 W8B6P2 A0A151IP62 A0A2A3ELA5
A0A194RHR5 A0A2A4J3B3 A0A2W1BP27 A0A212F8L3 A0A2H1VGH7 I4DNF7 A0A194PMC3 H9JNB3 A0A2A4KA95 A0A194QN55 A0A2A4K9Y1 A0A194PY14 A0A1J1IHN1 A0A1B0C9J9 A0A2A4K9Y5 A0A2H1W3H1 A0A1Q3FH65 V5IAH1 D6WKJ1 B0X4Q4 A0A1Y1KD57 A0A1Y1K9J5 Q17CS1 D6WKJ0 A0A088ALA3 A0A023EUG3 A0A182H4N7 A0A0L7QRN1 A0A182T1C6 H9JNI6 A0A154P8V9 A0A182PAB8 A0A182Y4D6 A0A2W1BCH7 A0A182VI17 A0A2A3ESR9 K7ITG9 A0A182TEQ3 A0A182QLT3 A0A2H1VUP3 A0A0M9A4G4 A0A182WRH0 Q7QDB3 A0A182HUA7 D6X4T8 A0A2M4CTR9 A0A2M4CU13 A0A182W090 A0A182K794 A0A310SN29 B3NK59 A0A2J7RBY0 B4HPT1 A0A194QK42 U4UII8 A0A0M9A1C2 N6T164 A0A182MBN7 A0A182JCT1 A0A182NFM5 Q7K3M6 B4PAQ3 B4QEI8 E2BIH0 A0A232F564 A0A182RNN4 A0A2A4J566 A0A182F7G4 A0A212FFR4 A0A2W1BI79 A0A195DRF1 A0A194QIQ7 B3MH68 A0A088A3V4 B4MRV4 A0A0M3QVP8 K7JAK3 A0A1W4VGV5 A0A084VXP9 A0A310SUT5 E2A1J5 A0A1B6EKM5 A0A026W714 A0A158NPS8 A0A195ATZ5 B4JWE6 A0A0L7LUK1 A0A195FCZ0 A0A232EL12 E1ZZY2 W8B6P2 A0A151IP62 A0A2A3ELA5
Pubmed
19121390
28756777
26227816
26354079
22118469
22651552
+ More
18362917 19820115 28004739 17510324 24945155 26483478 25244985 20075255 12364791 14747013 17210077 17994087 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20798317 28648823 24438588 24508170 21347285 24495485
18362917 19820115 28004739 17510324 24945155 26483478 25244985 20075255 12364791 14747013 17210077 17994087 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20798317 28648823 24438588 24508170 21347285 24495485
EMBL
BABH01044534
BABH01044535
BABH01034412
BABH01034413
ODYU01003353
SOQ42005.1
+ More
KZ150304 PZC71520.1 JTDY01000054 KOB79138.1 NWSH01000158 PCG78950.1 KQ460367 KPJ15486.1 NWSH01003717 PCG65892.1 KZ149948 PZC76718.1 AGBW02009713 OWR50049.1 ODYU01002452 SOQ39959.1 AK402884 BAM19447.1 KQ459600 KPI94138.1 BABH01019600 NWSH01000020 PCG80713.1 KQ461195 KPJ06789.1 PCG80714.1 KQ459589 KPI97639.1 CVRI01000052 CRK99712.1 AJWK01002434 AJWK01002435 AJWK01002436 PCG80708.1 ODYU01006026 SOQ47506.1 GFDL01008134 JAV26911.1 GALX01001195 JAB67271.1 KQ971342 EFA04007.2 DS232350 EDS40471.1 GEZM01088532 JAV58130.1 GEZM01088533 JAV58129.1 CH477305 EAT44124.1 EFA04006.1 GAPW01001011 JAC12587.1 JXUM01109993 KQ565372 KXJ71040.1 KQ414775 KOC61282.1 BABH01019597 KQ434827 KZC07658.1 KZ150301 PZC71534.1 KZ288189 PBC34564.1 AAZX01006832 AXCN02000238 ODYU01004551 SOQ44555.1 KQ435759 KOX75743.1 AAAB01008859 EAA07935.5 APCN01000614 KQ971381 EEZ97586.2 GGFL01004566 MBW68744.1 GGFL01004591 MBW68769.1 KQ762190 OAD56165.1 CH954179 EDV55264.1 NEVH01005889 PNF38331.1 CH480816 EDW48649.1 KQ461198 KPJ05943.1 KB632314 ERL92208.1 KQ435794 KOX73779.1 APGK01048466 KB741110 ENN73834.1 AXCM01003853 AE013599 AY058478 BT120184 AAF57511.1 AAL13707.1 ADB91432.1 CM000158 EDW91444.1 KRJ99843.1 CM000362 CM002911 EDX07862.1 KMY95206.1 GL448504 EFN84558.1 NNAY01000983 OXU25618.1 NWSH01003004 PCG67121.1 AGBW02008774 OWR52572.1 KZ150029 PZC74769.1 KQ980581 KYN15387.1 KQ459185 KPJ03336.1 CH902619 EDV36845.1 CH963850 EDW74843.1 CP012524 ALC42779.1 AAZX01002446 ATLV01018145 KE525215 KFB42743.1 KQ760085 OAD61975.1 GL435766 EFN72668.1 GECZ01031285 JAS38484.1 KK107372 EZA51773.1 ADTU01022726 KQ976745 KYM75454.1 CH916375 EDV98284.1 KOB79137.1 KQ981673 KYN38256.1 NNAY01003667 OXU19035.1 GL435531 EFN73254.1 GAMC01009685 JAB96870.1 KQ976914 KYN07128.1 KZ288225 PBC31939.1
KZ150304 PZC71520.1 JTDY01000054 KOB79138.1 NWSH01000158 PCG78950.1 KQ460367 KPJ15486.1 NWSH01003717 PCG65892.1 KZ149948 PZC76718.1 AGBW02009713 OWR50049.1 ODYU01002452 SOQ39959.1 AK402884 BAM19447.1 KQ459600 KPI94138.1 BABH01019600 NWSH01000020 PCG80713.1 KQ461195 KPJ06789.1 PCG80714.1 KQ459589 KPI97639.1 CVRI01000052 CRK99712.1 AJWK01002434 AJWK01002435 AJWK01002436 PCG80708.1 ODYU01006026 SOQ47506.1 GFDL01008134 JAV26911.1 GALX01001195 JAB67271.1 KQ971342 EFA04007.2 DS232350 EDS40471.1 GEZM01088532 JAV58130.1 GEZM01088533 JAV58129.1 CH477305 EAT44124.1 EFA04006.1 GAPW01001011 JAC12587.1 JXUM01109993 KQ565372 KXJ71040.1 KQ414775 KOC61282.1 BABH01019597 KQ434827 KZC07658.1 KZ150301 PZC71534.1 KZ288189 PBC34564.1 AAZX01006832 AXCN02000238 ODYU01004551 SOQ44555.1 KQ435759 KOX75743.1 AAAB01008859 EAA07935.5 APCN01000614 KQ971381 EEZ97586.2 GGFL01004566 MBW68744.1 GGFL01004591 MBW68769.1 KQ762190 OAD56165.1 CH954179 EDV55264.1 NEVH01005889 PNF38331.1 CH480816 EDW48649.1 KQ461198 KPJ05943.1 KB632314 ERL92208.1 KQ435794 KOX73779.1 APGK01048466 KB741110 ENN73834.1 AXCM01003853 AE013599 AY058478 BT120184 AAF57511.1 AAL13707.1 ADB91432.1 CM000158 EDW91444.1 KRJ99843.1 CM000362 CM002911 EDX07862.1 KMY95206.1 GL448504 EFN84558.1 NNAY01000983 OXU25618.1 NWSH01003004 PCG67121.1 AGBW02008774 OWR52572.1 KZ150029 PZC74769.1 KQ980581 KYN15387.1 KQ459185 KPJ03336.1 CH902619 EDV36845.1 CH963850 EDW74843.1 CP012524 ALC42779.1 AAZX01002446 ATLV01018145 KE525215 KFB42743.1 KQ760085 OAD61975.1 GL435766 EFN72668.1 GECZ01031285 JAS38484.1 KK107372 EZA51773.1 ADTU01022726 KQ976745 KYM75454.1 CH916375 EDV98284.1 KOB79137.1 KQ981673 KYN38256.1 NNAY01003667 OXU19035.1 GL435531 EFN73254.1 GAMC01009685 JAB96870.1 KQ976914 KYN07128.1 KZ288225 PBC31939.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000007151
UP000053268
+ More
UP000183832 UP000092461 UP000007266 UP000002320 UP000008820 UP000005203 UP000069940 UP000249989 UP000053825 UP000075901 UP000076502 UP000075885 UP000076408 UP000075903 UP000242457 UP000002358 UP000075902 UP000075886 UP000053105 UP000076407 UP000007062 UP000075840 UP000075920 UP000075881 UP000008711 UP000235965 UP000001292 UP000030742 UP000019118 UP000075883 UP000075880 UP000075884 UP000000803 UP000002282 UP000000304 UP000008237 UP000215335 UP000075900 UP000069272 UP000078492 UP000007801 UP000007798 UP000092553 UP000192221 UP000030765 UP000000311 UP000053097 UP000005205 UP000078540 UP000001070 UP000078541 UP000078542
UP000183832 UP000092461 UP000007266 UP000002320 UP000008820 UP000005203 UP000069940 UP000249989 UP000053825 UP000075901 UP000076502 UP000075885 UP000076408 UP000075903 UP000242457 UP000002358 UP000075902 UP000075886 UP000053105 UP000076407 UP000007062 UP000075840 UP000075920 UP000075881 UP000008711 UP000235965 UP000001292 UP000030742 UP000019118 UP000075883 UP000075880 UP000075884 UP000000803 UP000002282 UP000000304 UP000008237 UP000215335 UP000075900 UP000069272 UP000078492 UP000007801 UP000007798 UP000092553 UP000192221 UP000030765 UP000000311 UP000053097 UP000005205 UP000078540 UP000001070 UP000078541 UP000078542
Pfam
Interpro
IPR020846
MFS_dom
+ More
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR005829 Sugar_transporter_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR022100 Mcl1_mid
IPR017986 WD40_repeat_dom
IPR019775 WD40_repeat_CS
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR024977 Apc4_WD40_dom
IPR011041 Quinoprot_gluc/sorb_DH
IPR015063 USP8_dimer
IPR011701 MFS
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR005829 Sugar_transporter_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR022100 Mcl1_mid
IPR017986 WD40_repeat_dom
IPR019775 WD40_repeat_CS
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR024977 Apc4_WD40_dom
IPR011041 Quinoprot_gluc/sorb_DH
IPR015063 USP8_dimer
IPR011701 MFS
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
Gene 3D
CDD
ProteinModelPortal
H9ISF5
H9JGL2
A0A2H1VMD7
A0A2W1BFH6
A0A0L7LUI9
A0A2A4K5L0
+ More
A0A194RHR5 A0A2A4J3B3 A0A2W1BP27 A0A212F8L3 A0A2H1VGH7 I4DNF7 A0A194PMC3 H9JNB3 A0A2A4KA95 A0A194QN55 A0A2A4K9Y1 A0A194PY14 A0A1J1IHN1 A0A1B0C9J9 A0A2A4K9Y5 A0A2H1W3H1 A0A1Q3FH65 V5IAH1 D6WKJ1 B0X4Q4 A0A1Y1KD57 A0A1Y1K9J5 Q17CS1 D6WKJ0 A0A088ALA3 A0A023EUG3 A0A182H4N7 A0A0L7QRN1 A0A182T1C6 H9JNI6 A0A154P8V9 A0A182PAB8 A0A182Y4D6 A0A2W1BCH7 A0A182VI17 A0A2A3ESR9 K7ITG9 A0A182TEQ3 A0A182QLT3 A0A2H1VUP3 A0A0M9A4G4 A0A182WRH0 Q7QDB3 A0A182HUA7 D6X4T8 A0A2M4CTR9 A0A2M4CU13 A0A182W090 A0A182K794 A0A310SN29 B3NK59 A0A2J7RBY0 B4HPT1 A0A194QK42 U4UII8 A0A0M9A1C2 N6T164 A0A182MBN7 A0A182JCT1 A0A182NFM5 Q7K3M6 B4PAQ3 B4QEI8 E2BIH0 A0A232F564 A0A182RNN4 A0A2A4J566 A0A182F7G4 A0A212FFR4 A0A2W1BI79 A0A195DRF1 A0A194QIQ7 B3MH68 A0A088A3V4 B4MRV4 A0A0M3QVP8 K7JAK3 A0A1W4VGV5 A0A084VXP9 A0A310SUT5 E2A1J5 A0A1B6EKM5 A0A026W714 A0A158NPS8 A0A195ATZ5 B4JWE6 A0A0L7LUK1 A0A195FCZ0 A0A232EL12 E1ZZY2 W8B6P2 A0A151IP62 A0A2A3ELA5
A0A194RHR5 A0A2A4J3B3 A0A2W1BP27 A0A212F8L3 A0A2H1VGH7 I4DNF7 A0A194PMC3 H9JNB3 A0A2A4KA95 A0A194QN55 A0A2A4K9Y1 A0A194PY14 A0A1J1IHN1 A0A1B0C9J9 A0A2A4K9Y5 A0A2H1W3H1 A0A1Q3FH65 V5IAH1 D6WKJ1 B0X4Q4 A0A1Y1KD57 A0A1Y1K9J5 Q17CS1 D6WKJ0 A0A088ALA3 A0A023EUG3 A0A182H4N7 A0A0L7QRN1 A0A182T1C6 H9JNI6 A0A154P8V9 A0A182PAB8 A0A182Y4D6 A0A2W1BCH7 A0A182VI17 A0A2A3ESR9 K7ITG9 A0A182TEQ3 A0A182QLT3 A0A2H1VUP3 A0A0M9A4G4 A0A182WRH0 Q7QDB3 A0A182HUA7 D6X4T8 A0A2M4CTR9 A0A2M4CU13 A0A182W090 A0A182K794 A0A310SN29 B3NK59 A0A2J7RBY0 B4HPT1 A0A194QK42 U4UII8 A0A0M9A1C2 N6T164 A0A182MBN7 A0A182JCT1 A0A182NFM5 Q7K3M6 B4PAQ3 B4QEI8 E2BIH0 A0A232F564 A0A182RNN4 A0A2A4J566 A0A182F7G4 A0A212FFR4 A0A2W1BI79 A0A195DRF1 A0A194QIQ7 B3MH68 A0A088A3V4 B4MRV4 A0A0M3QVP8 K7JAK3 A0A1W4VGV5 A0A084VXP9 A0A310SUT5 E2A1J5 A0A1B6EKM5 A0A026W714 A0A158NPS8 A0A195ATZ5 B4JWE6 A0A0L7LUK1 A0A195FCZ0 A0A232EL12 E1ZZY2 W8B6P2 A0A151IP62 A0A2A3ELA5
PDB
4QIQ
E-value=1.79811e-09,
Score=150
Ontologies
GO
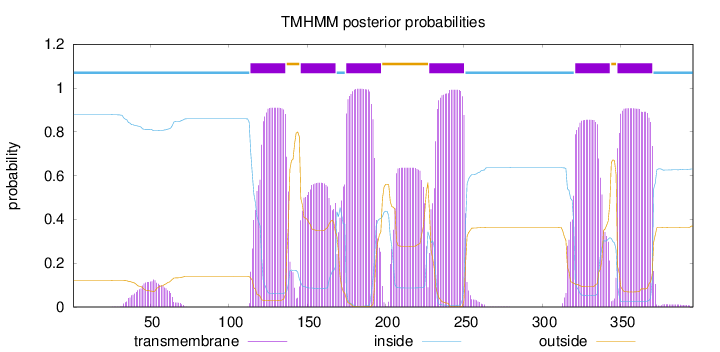
Topology
Length:
396
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
128.83242
Exp number, first 60 AAs:
2.34326
Total prob of N-in:
0.87841
inside
1 - 113
TMhelix
114 - 136
outside
137 - 145
TMhelix
146 - 168
inside
169 - 174
TMhelix
175 - 197
outside
198 - 227
TMhelix
228 - 250
inside
251 - 320
TMhelix
321 - 343
outside
344 - 347
TMhelix
348 - 370
inside
371 - 396
Population Genetic Test Statistics
Pi
232.376667
Theta
41.211669
Tajima's D
-1.549115
CLR
130.75318
CSRT
0.056247187640618
Interpretation
Uncertain
