Gene
KWMTBOMO04180
Pre Gene Modal
BGIBMGA008727
Annotation
PREDICTED:_nicotinamidase_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.461
Sequence
CDS
ATGTCGGAAGTTTCCAACGGCGTGGAATTCAACGAAGAAGTCTTCACTGAAATGGACGCTTGTTACGAAACTTTCGACAAAGACGGGGACGGAAAGCTGAATCTGGATGAGTTCCGGCTGATCTGCAAGGCGTTGTTCCGCAACGACAAGGGCCACATCTACCCGCTCTCCGAAGACAGAGCCAGACATATTTTCAACGTTTTCGATACAAACGGGGACGGGTTCATTGATAAGGAAGAGTTCACTTTCTGCTGGAACCATTGGATCAAGGTGATCGTGCGTCCGGTGAGCGCGTTCCTGATCGTGGACGTTCAGAACGACTTCATCTCCGGCACCCTCAACATCAGCAAGTGCAACGCCCAGCAAGACGGCAGCGAGGTGGTCGAACCCATAAACCATCTCCTGGATACGGTGCCGTTCGACTGTGTCTTCTACTCCCTGGACTGGCATCCCCCGGATCACGTGTCTTTCATCGACAACGTCCATATGAGAGAACTGCATCCATCCAGCCCGGTGTCGGCGGACAGGGCGGCCGCCTACGACACGGTGATCTTCGCCGGCCCCCCGCCCATGAGGCAGCGCCTTTGGCCGCGCCACTGCGTGCAGGACACGTGGGGCGCCGAGCTGCACAACGACCTCAAGATAGTCGAGGGCGCCACGAAAGTGTACAAGGGCACCAACCCGGAGGTGGACTCGTACTCCGTGTTCTGGGACAACAAGAAGCTGACCGACACCAGCCTCAGCACGCAGCTGCGACTGCGCGGCGCCACCGACATCTACATCTGCGGGCTGGCGTACGACGTGTGCGTCGGTGCAACGATAGCTGACGCGCTGGCCATCGGCTACCGCGCCGTGCTGGTGGAGGACGCCAGCCGGGGGGTCGACCTCGCTGACATCGAGAAGACGAAAGCAACCATCGTCAACAACAACGGAGTCATCGTGCGCTCTGACGAGGTGTTACCGATGGTGGAAGGGAGGGACCGGCGGCCCGAACTGGGCTACAAGCTCGCCATGGAACTGAAGAGGGCTATGGAGGAAAAATAA
Protein
MSEVSNGVEFNEEVFTEMDACYETFDKDGDGKLNLDEFRLICKALFRNDKGHIYPLSEDRARHIFNVFDTNGDGFIDKEEFTFCWNHWIKVIVRPVSAFLIVDVQNDFISGTLNISKCNAQQDGSEVVEPINHLLDTVPFDCVFYSLDWHPPDHVSFIDNVHMRELHPSSPVSADRAAAYDTVIFAGPPPMRQRLWPRHCVQDTWGAELHNDLKIVEGATKVYKGTNPEVDSYSVFWDNKKLTDTSLSTQLRLRGATDIYICGLAYDVCVGATIADALAIGYRAVLVEDASRGVDLADIEKTKATIVNNNGVIVRSDEVLPMVEGRDRRPELGYKLAMELKRAMEEK
Summary
Uniprot
H9JGT0
A0A2A4K4W5
A0A194PWE4
A0A212F532
A0A194RDF2
A0A1Y1KQX6
+ More
N6TQE5 A0A1W4WQC7 A0A182Y595 A0A2M4CGB5 A0A2M4CFV0 Q7PXJ7 A0A2M4AEB5 A0A2M4AC11 A0A182IWP9 A0A1Y9GZ86 A0A182N478 A0A1Y9GZ87 A0A1Q3FV39 A0A1Q3FUZ8 A0A2J7QFD8 U5EVX1 A0A1B0CJ37 A0A067R2Y1 T1PF57 A0A336MRR7 A0A0L0C929 K7J3G1 A0A1I8M3U9 B4JT25 A0A1B0FRG2 A0A1I8PQG0 A0A1L8E382 A0A1W4WRC9 B4NF00 A0A1W4WQD3 A0A1W4WFE6 A0A0A1WR36 A0A195FF67 A0A195CI31 B4K4T5 A0A0M4EJA8 E2BVX6 B4M4Q4 A0A0K8UXL0 A0A0V0G4A3 A0A195B783 A0A182TIJ0 A0A182HKW1 A0A1B6HXG8 A0A182X9Y0 A0A182RPZ0 A0A0C9QX80 A0A069DST2 B3LXR0 A0A023F4D7 A0A023F4C6 A0A1B6CXZ5 A0A182F744 A0A1W4W8H2 A0A182VYU7 A0A3B0K6N8 A0A151XAK2 A0A1B6M159 W8BQ83 Q17GI8 A0A026W375 E0VNM9 A0A182L980 B5DXP7 Q9VDU7 A0A310SKU7 A0A088A5G6 A0A0N0BJL6 A0A2A3EFQ4 A0A0L7R565 A0A182QZT8 A0A0J7KZ02 A0A158NG79 E2A8N4 B4PNJ2 B3P091 E9IUC4 A0A0P4VR18 A0A3L8DN74 A0A182P2C7 A0A1J1IDS2 J9JKX1 A0A2H8TRB4 A0A154PPV2 B4QSV1 A0A2S2R3J8 B4IHS9 T1IA36 A0A0A9XRM6 A0A0K8TIM5 D6W7R5 B4GLY3 V5GW71 A0A1A9WPT5
N6TQE5 A0A1W4WQC7 A0A182Y595 A0A2M4CGB5 A0A2M4CFV0 Q7PXJ7 A0A2M4AEB5 A0A2M4AC11 A0A182IWP9 A0A1Y9GZ86 A0A182N478 A0A1Y9GZ87 A0A1Q3FV39 A0A1Q3FUZ8 A0A2J7QFD8 U5EVX1 A0A1B0CJ37 A0A067R2Y1 T1PF57 A0A336MRR7 A0A0L0C929 K7J3G1 A0A1I8M3U9 B4JT25 A0A1B0FRG2 A0A1I8PQG0 A0A1L8E382 A0A1W4WRC9 B4NF00 A0A1W4WQD3 A0A1W4WFE6 A0A0A1WR36 A0A195FF67 A0A195CI31 B4K4T5 A0A0M4EJA8 E2BVX6 B4M4Q4 A0A0K8UXL0 A0A0V0G4A3 A0A195B783 A0A182TIJ0 A0A182HKW1 A0A1B6HXG8 A0A182X9Y0 A0A182RPZ0 A0A0C9QX80 A0A069DST2 B3LXR0 A0A023F4D7 A0A023F4C6 A0A1B6CXZ5 A0A182F744 A0A1W4W8H2 A0A182VYU7 A0A3B0K6N8 A0A151XAK2 A0A1B6M159 W8BQ83 Q17GI8 A0A026W375 E0VNM9 A0A182L980 B5DXP7 Q9VDU7 A0A310SKU7 A0A088A5G6 A0A0N0BJL6 A0A2A3EFQ4 A0A0L7R565 A0A182QZT8 A0A0J7KZ02 A0A158NG79 E2A8N4 B4PNJ2 B3P091 E9IUC4 A0A0P4VR18 A0A3L8DN74 A0A182P2C7 A0A1J1IDS2 J9JKX1 A0A2H8TRB4 A0A154PPV2 B4QSV1 A0A2S2R3J8 B4IHS9 T1IA36 A0A0A9XRM6 A0A0K8TIM5 D6W7R5 B4GLY3 V5GW71 A0A1A9WPT5
Pubmed
19121390
26354079
22118469
28004739
23537049
25244985
+ More
12364791 14747013 17210077 24845553 26108605 20075255 25315136 17994087 25830018 18057021 20798317 26334808 25474469 24495485 17510324 24508170 20566863 20966253 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 17550304 21282665 27129103 30249741 25401762 26823975 18362917 19820115
12364791 14747013 17210077 24845553 26108605 20075255 25315136 17994087 25830018 18057021 20798317 26334808 25474469 24495485 17510324 24508170 20566863 20966253 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 17550304 21282665 27129103 30249741 25401762 26823975 18362917 19820115
EMBL
BABH01034400
BABH01034401
NWSH01000158
PCG78954.1
KQ459589
KPI97647.1
+ More
AGBW02010265 OWR48845.1 KQ460367 KPJ15494.1 GEZM01078914 JAV62580.1 APGK01024787 KB740562 KB632399 ENN80273.1 ERL94823.1 GGFL01000013 MBW64191.1 GGFL01000015 MBW64193.1 AAAB01008987 EAA01751.5 GGFK01005769 MBW39090.1 GGFK01005005 MBW38326.1 GFDL01003647 JAV31398.1 GFDL01003670 JAV31375.1 NEVH01015299 PNF27305.1 GANO01001688 JAB58183.1 AJWK01014012 AJWK01014013 AJWK01014014 AJWK01014015 AJWK01014016 KK852747 KDR17274.1 KA646795 AFP61424.1 UFQT01001651 UFQT01001782 SSX31298.1 SSX31789.1 JRES01000752 KNC28760.1 CH916373 EDV94915.1 CCAG010001353 CCAG010001354 CCAG010001355 CCAG010001356 GFDF01000990 JAV13094.1 CH964251 EDW83375.2 GBXI01012768 JAD01524.1 KQ981625 KYN39006.1 KQ977721 KYN00391.1 CH933806 EDW16088.1 KRG01920.1 CP012526 ALC46946.1 GL451066 EFN80154.1 CH940652 EDW59615.1 KRF78963.1 GDHF01022166 GDHF01020993 JAI30148.1 JAI31321.1 GECL01003273 JAP02851.1 KQ976574 KYM80356.1 APCN01000873 GECU01028315 JAS79391.1 GBYB01000269 GBYB01000270 JAG70036.1 JAG70037.1 GBGD01001889 JAC87000.1 CH902617 EDV41717.2 KPU79309.1 GBBI01002336 JAC16376.1 GBBI01002337 JAC16375.1 GEDC01019093 GEDC01015604 JAS18205.1 JAS21694.1 OUUW01000008 SPP83730.1 KQ982339 KYQ57394.1 GEBQ01017328 GEBQ01010353 JAT22649.1 JAT29624.1 GAMC01005368 JAC01188.1 CH477259 EAT45737.1 KK107487 EZA50036.1 DS235341 EEB14985.1 CM000070 EDY68647.4 AE014297 AY122151 AAF55693.3 AAM52663.1 AGB96118.1 KQ761783 OAD56989.1 KQ435715 KOX79091.1 KZ288256 PBC30548.1 KQ414654 KOC65964.1 AXCN02000028 LBMM01001939 KMQ95509.1 ADTU01015041 GL437652 EFN70221.1 CM000160 EDW96055.1 KRK02868.1 KRK02869.1 KRK02870.1 CH954181 EDV48465.1 GL765940 EFZ15829.1 GDKW01001125 JAI55470.1 QOIP01000006 RLU21662.1 CVRI01000047 CRK97700.1 ABLF02034749 ABLF02034750 GFXV01004918 MBW16723.1 KQ434998 KZC13278.1 CM000364 EDX12311.1 GGMS01015408 MBY84611.1 CH480840 EDW49446.1 ACPB03012045 GBHO01023879 GBHO01023877 GBHO01023876 GBHO01023875 GBHO01007154 JAG19725.1 JAG19727.1 JAG19728.1 JAG19729.1 JAG36450.1 GBRD01000467 GDHC01021033 JAG65354.1 JAP97595.1 KQ971307 EFA11335.1 CH479185 EDW38557.1 GALX01003998 JAB64468.1
AGBW02010265 OWR48845.1 KQ460367 KPJ15494.1 GEZM01078914 JAV62580.1 APGK01024787 KB740562 KB632399 ENN80273.1 ERL94823.1 GGFL01000013 MBW64191.1 GGFL01000015 MBW64193.1 AAAB01008987 EAA01751.5 GGFK01005769 MBW39090.1 GGFK01005005 MBW38326.1 GFDL01003647 JAV31398.1 GFDL01003670 JAV31375.1 NEVH01015299 PNF27305.1 GANO01001688 JAB58183.1 AJWK01014012 AJWK01014013 AJWK01014014 AJWK01014015 AJWK01014016 KK852747 KDR17274.1 KA646795 AFP61424.1 UFQT01001651 UFQT01001782 SSX31298.1 SSX31789.1 JRES01000752 KNC28760.1 CH916373 EDV94915.1 CCAG010001353 CCAG010001354 CCAG010001355 CCAG010001356 GFDF01000990 JAV13094.1 CH964251 EDW83375.2 GBXI01012768 JAD01524.1 KQ981625 KYN39006.1 KQ977721 KYN00391.1 CH933806 EDW16088.1 KRG01920.1 CP012526 ALC46946.1 GL451066 EFN80154.1 CH940652 EDW59615.1 KRF78963.1 GDHF01022166 GDHF01020993 JAI30148.1 JAI31321.1 GECL01003273 JAP02851.1 KQ976574 KYM80356.1 APCN01000873 GECU01028315 JAS79391.1 GBYB01000269 GBYB01000270 JAG70036.1 JAG70037.1 GBGD01001889 JAC87000.1 CH902617 EDV41717.2 KPU79309.1 GBBI01002336 JAC16376.1 GBBI01002337 JAC16375.1 GEDC01019093 GEDC01015604 JAS18205.1 JAS21694.1 OUUW01000008 SPP83730.1 KQ982339 KYQ57394.1 GEBQ01017328 GEBQ01010353 JAT22649.1 JAT29624.1 GAMC01005368 JAC01188.1 CH477259 EAT45737.1 KK107487 EZA50036.1 DS235341 EEB14985.1 CM000070 EDY68647.4 AE014297 AY122151 AAF55693.3 AAM52663.1 AGB96118.1 KQ761783 OAD56989.1 KQ435715 KOX79091.1 KZ288256 PBC30548.1 KQ414654 KOC65964.1 AXCN02000028 LBMM01001939 KMQ95509.1 ADTU01015041 GL437652 EFN70221.1 CM000160 EDW96055.1 KRK02868.1 KRK02869.1 KRK02870.1 CH954181 EDV48465.1 GL765940 EFZ15829.1 GDKW01001125 JAI55470.1 QOIP01000006 RLU21662.1 CVRI01000047 CRK97700.1 ABLF02034749 ABLF02034750 GFXV01004918 MBW16723.1 KQ434998 KZC13278.1 CM000364 EDX12311.1 GGMS01015408 MBY84611.1 CH480840 EDW49446.1 ACPB03012045 GBHO01023879 GBHO01023877 GBHO01023876 GBHO01023875 GBHO01007154 JAG19725.1 JAG19727.1 JAG19728.1 JAG19729.1 JAG36450.1 GBRD01000467 GDHC01021033 JAG65354.1 JAP97595.1 KQ971307 EFA11335.1 CH479185 EDW38557.1 GALX01003998 JAB64468.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000019118
+ More
UP000030742 UP000192223 UP000076408 UP000007062 UP000075880 UP000075884 UP000235965 UP000092461 UP000027135 UP000037069 UP000002358 UP000095301 UP000001070 UP000092444 UP000095300 UP000007798 UP000078541 UP000078542 UP000009192 UP000092553 UP000008237 UP000008792 UP000078540 UP000075902 UP000075840 UP000076407 UP000075900 UP000007801 UP000069272 UP000192221 UP000075920 UP000268350 UP000075809 UP000008820 UP000053097 UP000009046 UP000075882 UP000001819 UP000000803 UP000005203 UP000053105 UP000242457 UP000053825 UP000075886 UP000036403 UP000005205 UP000000311 UP000002282 UP000008711 UP000279307 UP000075885 UP000183832 UP000007819 UP000076502 UP000000304 UP000001292 UP000015103 UP000007266 UP000008744 UP000091820
UP000030742 UP000192223 UP000076408 UP000007062 UP000075880 UP000075884 UP000235965 UP000092461 UP000027135 UP000037069 UP000002358 UP000095301 UP000001070 UP000092444 UP000095300 UP000007798 UP000078541 UP000078542 UP000009192 UP000092553 UP000008237 UP000008792 UP000078540 UP000075902 UP000075840 UP000076407 UP000075900 UP000007801 UP000069272 UP000192221 UP000075920 UP000268350 UP000075809 UP000008820 UP000053097 UP000009046 UP000075882 UP000001819 UP000000803 UP000005203 UP000053105 UP000242457 UP000053825 UP000075886 UP000036403 UP000005205 UP000000311 UP000002282 UP000008711 UP000279307 UP000075885 UP000183832 UP000007819 UP000076502 UP000000304 UP000001292 UP000015103 UP000007266 UP000008744 UP000091820
Pfam
Interpro
IPR011992
EF-hand-dom_pair
+ More
IPR000868 Isochorismatase-like
IPR002048 EF_hand_dom
IPR036380 Isochorismatase-like_sf
IPR018247 EF_Hand_1_Ca_BS
IPR036612 KH_dom_type_1_sf
IPR012340 NA-bd_OB-fold
IPR025721 Exosome_cplx_N_dom
IPR022967 S1_dom
IPR004088 KH_dom_type_1
IPR026699 Exosome_RNA_bind1/RRP40/RRP4
IPR003029 S1_domain
IPR000868 Isochorismatase-like
IPR002048 EF_hand_dom
IPR036380 Isochorismatase-like_sf
IPR018247 EF_Hand_1_Ca_BS
IPR036612 KH_dom_type_1_sf
IPR012340 NA-bd_OB-fold
IPR025721 Exosome_cplx_N_dom
IPR022967 S1_dom
IPR004088 KH_dom_type_1
IPR026699 Exosome_RNA_bind1/RRP40/RRP4
IPR003029 S1_domain
Gene 3D
CDD
ProteinModelPortal
H9JGT0
A0A2A4K4W5
A0A194PWE4
A0A212F532
A0A194RDF2
A0A1Y1KQX6
+ More
N6TQE5 A0A1W4WQC7 A0A182Y595 A0A2M4CGB5 A0A2M4CFV0 Q7PXJ7 A0A2M4AEB5 A0A2M4AC11 A0A182IWP9 A0A1Y9GZ86 A0A182N478 A0A1Y9GZ87 A0A1Q3FV39 A0A1Q3FUZ8 A0A2J7QFD8 U5EVX1 A0A1B0CJ37 A0A067R2Y1 T1PF57 A0A336MRR7 A0A0L0C929 K7J3G1 A0A1I8M3U9 B4JT25 A0A1B0FRG2 A0A1I8PQG0 A0A1L8E382 A0A1W4WRC9 B4NF00 A0A1W4WQD3 A0A1W4WFE6 A0A0A1WR36 A0A195FF67 A0A195CI31 B4K4T5 A0A0M4EJA8 E2BVX6 B4M4Q4 A0A0K8UXL0 A0A0V0G4A3 A0A195B783 A0A182TIJ0 A0A182HKW1 A0A1B6HXG8 A0A182X9Y0 A0A182RPZ0 A0A0C9QX80 A0A069DST2 B3LXR0 A0A023F4D7 A0A023F4C6 A0A1B6CXZ5 A0A182F744 A0A1W4W8H2 A0A182VYU7 A0A3B0K6N8 A0A151XAK2 A0A1B6M159 W8BQ83 Q17GI8 A0A026W375 E0VNM9 A0A182L980 B5DXP7 Q9VDU7 A0A310SKU7 A0A088A5G6 A0A0N0BJL6 A0A2A3EFQ4 A0A0L7R565 A0A182QZT8 A0A0J7KZ02 A0A158NG79 E2A8N4 B4PNJ2 B3P091 E9IUC4 A0A0P4VR18 A0A3L8DN74 A0A182P2C7 A0A1J1IDS2 J9JKX1 A0A2H8TRB4 A0A154PPV2 B4QSV1 A0A2S2R3J8 B4IHS9 T1IA36 A0A0A9XRM6 A0A0K8TIM5 D6W7R5 B4GLY3 V5GW71 A0A1A9WPT5
N6TQE5 A0A1W4WQC7 A0A182Y595 A0A2M4CGB5 A0A2M4CFV0 Q7PXJ7 A0A2M4AEB5 A0A2M4AC11 A0A182IWP9 A0A1Y9GZ86 A0A182N478 A0A1Y9GZ87 A0A1Q3FV39 A0A1Q3FUZ8 A0A2J7QFD8 U5EVX1 A0A1B0CJ37 A0A067R2Y1 T1PF57 A0A336MRR7 A0A0L0C929 K7J3G1 A0A1I8M3U9 B4JT25 A0A1B0FRG2 A0A1I8PQG0 A0A1L8E382 A0A1W4WRC9 B4NF00 A0A1W4WQD3 A0A1W4WFE6 A0A0A1WR36 A0A195FF67 A0A195CI31 B4K4T5 A0A0M4EJA8 E2BVX6 B4M4Q4 A0A0K8UXL0 A0A0V0G4A3 A0A195B783 A0A182TIJ0 A0A182HKW1 A0A1B6HXG8 A0A182X9Y0 A0A182RPZ0 A0A0C9QX80 A0A069DST2 B3LXR0 A0A023F4D7 A0A023F4C6 A0A1B6CXZ5 A0A182F744 A0A1W4W8H2 A0A182VYU7 A0A3B0K6N8 A0A151XAK2 A0A1B6M159 W8BQ83 Q17GI8 A0A026W375 E0VNM9 A0A182L980 B5DXP7 Q9VDU7 A0A310SKU7 A0A088A5G6 A0A0N0BJL6 A0A2A3EFQ4 A0A0L7R565 A0A182QZT8 A0A0J7KZ02 A0A158NG79 E2A8N4 B4PNJ2 B3P091 E9IUC4 A0A0P4VR18 A0A3L8DN74 A0A182P2C7 A0A1J1IDS2 J9JKX1 A0A2H8TRB4 A0A154PPV2 B4QSV1 A0A2S2R3J8 B4IHS9 T1IA36 A0A0A9XRM6 A0A0K8TIM5 D6W7R5 B4GLY3 V5GW71 A0A1A9WPT5
PDB
2WTA
E-value=1.41404e-27,
Score=305
Ontologies
GO
PANTHER
Topology
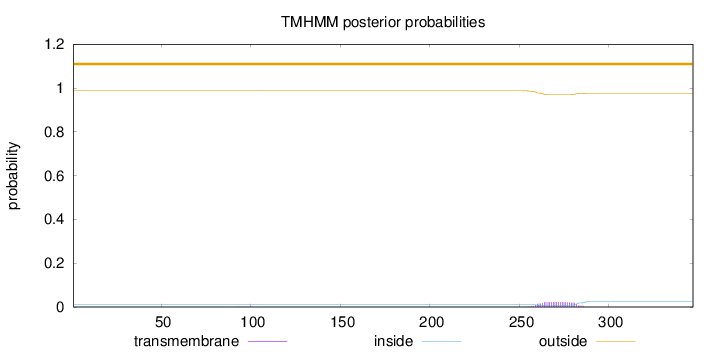
Length:
347
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.51432
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01266
outside
1 - 347
Population Genetic Test Statistics
Pi
243.009768
Theta
165.798052
Tajima's D
1.805107
CLR
0.314423
CSRT
0.847357632118394
Interpretation
Uncertain
