Gene
KWMTBOMO04177
Pre Gene Modal
BGIBMGA008665
Annotation
Chymotrypsinogen_B_[Operophtera_brumata]
Full name
Protein masquerade
Location in the cell
Extracellular Reliability : 2.087
Sequence
CDS
ATGCTTTTTCCAGCGGGTCCGATCCCTCTGAGGGTACGCGAGGCTGAAATCCCGATAGTAAGTGACGCCGAGTGCATCCGTAAAGTGAACGCAGTCACCGAGAAGATTTTCATCCTCCCCGCGAGTTCCTTCTGCGCGGGGGGGGAGGAAGGAAACGACGCCTGTCAGGGTGATGGTGGAGGACCACTAGTGTGTCAAGACGACGGGTTCTACGAGCTGGCGGGGCTGGTGTCCTGGGGCTTCGGCTGCGGCCGCAGGGACGTACCAGGGGTCTACGTTAAGGTCTCCGCCTTCATAGGGTGGATCAATCAGATAATATCCGTCAACAATAACAATAACAACATAGACTGA
Protein
MLFPAGPIPLRVREAEIPIVSDAECIRKVNAVTEKIFILPASSFCAGGEEGNDACQGDGGGPLVCQDDGFYELAGLVSWGFGCGRRDVPGVYVKVSAFIGWINQIISVNNNNNNID
Summary
Description
In embryogenesis, has a role in somatic muscle attachment and in the development of axonal pathways probably by stabilizing cell-matrix adhesion and/or by acting as a competitive antagonist of serine proteases.
Similarity
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family. CLIP subfamily.
Belongs to the peptidase S1 family. CLIP subfamily.
Keywords
Alternative splicing
Cell projection
Complete proteome
Developmental protein
Disulfide bond
Glycoprotein
Reference proteome
Repeat
Secreted
Serine protease homolog
Signal
Feature
chain Protein masquerade
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A1B6FVP8
A0A232FI05
K7J2Z9
A0A0M9ABM3
H9JGL8
A0A0L7LBM5
+ More
A0A0J7K5T0 A0A0L0CDQ7 A0A154PPQ2 A0A224XZ37 A0A034V031 A0A0L7RHX1 A0A0K8V7H3 A0A158NB79 A0A0A1XE51 A0A1A9YIH3 N6T8S3 A0A1I8Q9G9 W8BLB7 A0A1B0AKS2 U4UJF9 A0A067R6E8 A0A1A9WLK0 F4X698 A0A1A9UG58 A0A151IDY9 A0A1A9ZKD4 A0A1B0FDP0 B4HU10 A0A0A9VWZ3 A0A151X314 A0A1I8MPY4 A0A212F547 A0A182G1N0 B0WB60 Q17J66 T1GEJ5 A0A084W000 A0A182IWB1 W5JCQ1 A0A182KA83 A0A182FQS9 X1XHE5 A0A195FTP4 A0A182RNX5 A0A182XHQ6 A0A182W001 A0A182UMY6 A0A182LGH9 A0A182HTN2 A0A182LRX7 Q7QCV2 A0A182Y447 A0A182SKQ3 A0A1S3DSP4 A0A0M3QWE9 A0A151JCB9 A0A182TUV2 A0A2H8TX95 E9J5F6 E2BTC3 A0A336MLD1 A0A182Q6P9 I4DMM2 A0A182P272 E0VIF7 A0A336M1M4 A0A1W4W872 E1ZUU6 A0A182N776 A0A3B0JXJ3 A0A2J7R1K2 A0A3B0JPM7 B4LB91 Q9VZH2-2 Q9VZH2 B3NG45 B4QQG6 B4PHQ5 B4J1F0 B4L9D3 A0A1J1HYT9 A0A2W1BQR2 A0A0J9RP82 A0A3L8DLM6 Q29D77 B4N3K3 A0A026WTF4 B4H7V6 A0A146L7U5 A0A1V1G575 B3M4V9 A0A1S3DWF4 A0A2H8TJX9 A0A194Q2L6 T1HZE5 A0A2A4K3W1 A0A2S2R3Z0 A0A1W4X873 A0A2P8Z9E4
A0A0J7K5T0 A0A0L0CDQ7 A0A154PPQ2 A0A224XZ37 A0A034V031 A0A0L7RHX1 A0A0K8V7H3 A0A158NB79 A0A0A1XE51 A0A1A9YIH3 N6T8S3 A0A1I8Q9G9 W8BLB7 A0A1B0AKS2 U4UJF9 A0A067R6E8 A0A1A9WLK0 F4X698 A0A1A9UG58 A0A151IDY9 A0A1A9ZKD4 A0A1B0FDP0 B4HU10 A0A0A9VWZ3 A0A151X314 A0A1I8MPY4 A0A212F547 A0A182G1N0 B0WB60 Q17J66 T1GEJ5 A0A084W000 A0A182IWB1 W5JCQ1 A0A182KA83 A0A182FQS9 X1XHE5 A0A195FTP4 A0A182RNX5 A0A182XHQ6 A0A182W001 A0A182UMY6 A0A182LGH9 A0A182HTN2 A0A182LRX7 Q7QCV2 A0A182Y447 A0A182SKQ3 A0A1S3DSP4 A0A0M3QWE9 A0A151JCB9 A0A182TUV2 A0A2H8TX95 E9J5F6 E2BTC3 A0A336MLD1 A0A182Q6P9 I4DMM2 A0A182P272 E0VIF7 A0A336M1M4 A0A1W4W872 E1ZUU6 A0A182N776 A0A3B0JXJ3 A0A2J7R1K2 A0A3B0JPM7 B4LB91 Q9VZH2-2 Q9VZH2 B3NG45 B4QQG6 B4PHQ5 B4J1F0 B4L9D3 A0A1J1HYT9 A0A2W1BQR2 A0A0J9RP82 A0A3L8DLM6 Q29D77 B4N3K3 A0A026WTF4 B4H7V6 A0A146L7U5 A0A1V1G575 B3M4V9 A0A1S3DWF4 A0A2H8TJX9 A0A194Q2L6 T1HZE5 A0A2A4K3W1 A0A2S2R3Z0 A0A1W4X873 A0A2P8Z9E4
Pubmed
28648823
20075255
19121390
26227816
26108605
25348373
+ More
21347285 25830018 23537049 24495485 24845553 21719571 17994087 25401762 25315136 22118469 26483478 17510324 24438588 20920257 23761445 20966253 12364791 14747013 17210077 25244985 21282665 20798317 22651552 20566863 7851790 10731132 12537572 12537569 8817456 17550304 28756777 22936249 30249741 15632085 24508170 26823975 28410430 26354079 29403074
21347285 25830018 23537049 24495485 24845553 21719571 17994087 25401762 25315136 22118469 26483478 17510324 24438588 20920257 23761445 20966253 12364791 14747013 17210077 25244985 21282665 20798317 22651552 20566863 7851790 10731132 12537572 12537569 8817456 17550304 28756777 22936249 30249741 15632085 24508170 26823975 28410430 26354079 29403074
EMBL
GECZ01015483
JAS54286.1
NNAY01000174
OXU30312.1
KQ435710
KOX79829.1
+ More
BABH01034397 JTDY01001818 KOB72795.1 LBMM01013296 KMQ85717.1 JRES01000645 KNC29624.1 KQ435007 KZC13414.1 GFTR01003115 JAW13311.1 GAKP01022346 JAC36606.1 KQ414588 KOC70455.1 GDHF01017432 JAI34882.1 ADTU01010489 ADTU01010490 GBXI01005554 JAD08738.1 APGK01047061 APGK01047062 KB741077 ENN74098.1 GAMC01006983 GAMC01006982 JAB99572.1 JXJN01027882 KB632384 ERL94229.1 KK852669 KDR18860.1 GL888797 EGI58028.1 KQ977901 KYM98880.1 CCAG010020298 CH480817 EDW50431.1 GBHO01043450 JAG00154.1 KQ982577 KYQ54640.1 AGBW02010265 OWR48843.1 JXUM01137384 KQ568570 KXJ68952.1 DS231877 EDS42125.1 CH477234 EAT46741.2 CAQQ02391959 ATLV01019061 KE525259 KFB43544.1 ADMH02001605 ETN61856.1 ABLF02016015 KQ981272 KYN43833.1 APCN01000570 AXCM01001634 AAAB01008859 EAA08096.5 CP012525 ALC44029.1 KQ979060 KYN23168.1 GFXV01006337 MBW18142.1 GL768153 EFZ11946.1 GL450389 EFN81044.1 UFQT01001460 SSX30655.1 AXCN02000328 AK402540 BAM19162.1 DS235199 EEB13163.1 UFQT01000280 SSX22733.1 GL434297 EFN75038.1 OUUW01000002 SPP77441.1 NEVH01008206 PNF34708.1 SPP77440.1 CH940647 EDW68655.2 U18130 AE014296 AY052044 BT001597 AAC46512.1 AAF47850.1 AAK93468.1 AAN71352.1 CH954178 EDV50874.1 CM000363 EDX09172.1 CM000159 EDW93364.2 CH916366 EDV95841.1 CH933816 EDW17308.2 CVRI01000029 CRK92492.1 KZ149929 PZC77439.1 CM002912 KMY97517.1 QOIP01000006 RLU21281.1 CH379070 EAL30537.3 CH964095 EDW79208.2 KK107128 EZA58384.1 CH479219 EDW34746.1 GDHC01015333 JAQ03296.1 FX985333 BAX07346.1 CH902618 EDV40533.2 GFXV01001783 MBW13588.1 KQ459589 KPI97650.1 ACPB03014181 NWSH01000158 PCG78951.1 GGMS01015558 MBY84761.1 PYGN01000139 PSN53112.1
BABH01034397 JTDY01001818 KOB72795.1 LBMM01013296 KMQ85717.1 JRES01000645 KNC29624.1 KQ435007 KZC13414.1 GFTR01003115 JAW13311.1 GAKP01022346 JAC36606.1 KQ414588 KOC70455.1 GDHF01017432 JAI34882.1 ADTU01010489 ADTU01010490 GBXI01005554 JAD08738.1 APGK01047061 APGK01047062 KB741077 ENN74098.1 GAMC01006983 GAMC01006982 JAB99572.1 JXJN01027882 KB632384 ERL94229.1 KK852669 KDR18860.1 GL888797 EGI58028.1 KQ977901 KYM98880.1 CCAG010020298 CH480817 EDW50431.1 GBHO01043450 JAG00154.1 KQ982577 KYQ54640.1 AGBW02010265 OWR48843.1 JXUM01137384 KQ568570 KXJ68952.1 DS231877 EDS42125.1 CH477234 EAT46741.2 CAQQ02391959 ATLV01019061 KE525259 KFB43544.1 ADMH02001605 ETN61856.1 ABLF02016015 KQ981272 KYN43833.1 APCN01000570 AXCM01001634 AAAB01008859 EAA08096.5 CP012525 ALC44029.1 KQ979060 KYN23168.1 GFXV01006337 MBW18142.1 GL768153 EFZ11946.1 GL450389 EFN81044.1 UFQT01001460 SSX30655.1 AXCN02000328 AK402540 BAM19162.1 DS235199 EEB13163.1 UFQT01000280 SSX22733.1 GL434297 EFN75038.1 OUUW01000002 SPP77441.1 NEVH01008206 PNF34708.1 SPP77440.1 CH940647 EDW68655.2 U18130 AE014296 AY052044 BT001597 AAC46512.1 AAF47850.1 AAK93468.1 AAN71352.1 CH954178 EDV50874.1 CM000363 EDX09172.1 CM000159 EDW93364.2 CH916366 EDV95841.1 CH933816 EDW17308.2 CVRI01000029 CRK92492.1 KZ149929 PZC77439.1 CM002912 KMY97517.1 QOIP01000006 RLU21281.1 CH379070 EAL30537.3 CH964095 EDW79208.2 KK107128 EZA58384.1 CH479219 EDW34746.1 GDHC01015333 JAQ03296.1 FX985333 BAX07346.1 CH902618 EDV40533.2 GFXV01001783 MBW13588.1 KQ459589 KPI97650.1 ACPB03014181 NWSH01000158 PCG78951.1 GGMS01015558 MBY84761.1 PYGN01000139 PSN53112.1
Proteomes
UP000215335
UP000002358
UP000053105
UP000005204
UP000037510
UP000036403
+ More
UP000037069 UP000076502 UP000053825 UP000005205 UP000092443 UP000019118 UP000095300 UP000092460 UP000030742 UP000027135 UP000091820 UP000007755 UP000078200 UP000078542 UP000092445 UP000092444 UP000001292 UP000075809 UP000095301 UP000007151 UP000069940 UP000249989 UP000002320 UP000008820 UP000015102 UP000030765 UP000075880 UP000000673 UP000075881 UP000069272 UP000007819 UP000078541 UP000075900 UP000076407 UP000075920 UP000075903 UP000075882 UP000075840 UP000075883 UP000007062 UP000076408 UP000075901 UP000079169 UP000092553 UP000078492 UP000075902 UP000008237 UP000075886 UP000075885 UP000009046 UP000192221 UP000000311 UP000075884 UP000268350 UP000235965 UP000008792 UP000000803 UP000008711 UP000000304 UP000002282 UP000001070 UP000009192 UP000183832 UP000279307 UP000001819 UP000007798 UP000053097 UP000008744 UP000007801 UP000053268 UP000015103 UP000218220 UP000192223 UP000245037
UP000037069 UP000076502 UP000053825 UP000005205 UP000092443 UP000019118 UP000095300 UP000092460 UP000030742 UP000027135 UP000091820 UP000007755 UP000078200 UP000078542 UP000092445 UP000092444 UP000001292 UP000075809 UP000095301 UP000007151 UP000069940 UP000249989 UP000002320 UP000008820 UP000015102 UP000030765 UP000075880 UP000000673 UP000075881 UP000069272 UP000007819 UP000078541 UP000075900 UP000076407 UP000075920 UP000075903 UP000075882 UP000075840 UP000075883 UP000007062 UP000076408 UP000075901 UP000079169 UP000092553 UP000078492 UP000075902 UP000008237 UP000075886 UP000075885 UP000009046 UP000192221 UP000000311 UP000075884 UP000268350 UP000235965 UP000008792 UP000000803 UP000008711 UP000000304 UP000002282 UP000001070 UP000009192 UP000183832 UP000279307 UP000001819 UP000007798 UP000053097 UP000008744 UP000007801 UP000053268 UP000015103 UP000218220 UP000192223 UP000245037
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
A0A1B6FVP8
A0A232FI05
K7J2Z9
A0A0M9ABM3
H9JGL8
A0A0L7LBM5
+ More
A0A0J7K5T0 A0A0L0CDQ7 A0A154PPQ2 A0A224XZ37 A0A034V031 A0A0L7RHX1 A0A0K8V7H3 A0A158NB79 A0A0A1XE51 A0A1A9YIH3 N6T8S3 A0A1I8Q9G9 W8BLB7 A0A1B0AKS2 U4UJF9 A0A067R6E8 A0A1A9WLK0 F4X698 A0A1A9UG58 A0A151IDY9 A0A1A9ZKD4 A0A1B0FDP0 B4HU10 A0A0A9VWZ3 A0A151X314 A0A1I8MPY4 A0A212F547 A0A182G1N0 B0WB60 Q17J66 T1GEJ5 A0A084W000 A0A182IWB1 W5JCQ1 A0A182KA83 A0A182FQS9 X1XHE5 A0A195FTP4 A0A182RNX5 A0A182XHQ6 A0A182W001 A0A182UMY6 A0A182LGH9 A0A182HTN2 A0A182LRX7 Q7QCV2 A0A182Y447 A0A182SKQ3 A0A1S3DSP4 A0A0M3QWE9 A0A151JCB9 A0A182TUV2 A0A2H8TX95 E9J5F6 E2BTC3 A0A336MLD1 A0A182Q6P9 I4DMM2 A0A182P272 E0VIF7 A0A336M1M4 A0A1W4W872 E1ZUU6 A0A182N776 A0A3B0JXJ3 A0A2J7R1K2 A0A3B0JPM7 B4LB91 Q9VZH2-2 Q9VZH2 B3NG45 B4QQG6 B4PHQ5 B4J1F0 B4L9D3 A0A1J1HYT9 A0A2W1BQR2 A0A0J9RP82 A0A3L8DLM6 Q29D77 B4N3K3 A0A026WTF4 B4H7V6 A0A146L7U5 A0A1V1G575 B3M4V9 A0A1S3DWF4 A0A2H8TJX9 A0A194Q2L6 T1HZE5 A0A2A4K3W1 A0A2S2R3Z0 A0A1W4X873 A0A2P8Z9E4
A0A0J7K5T0 A0A0L0CDQ7 A0A154PPQ2 A0A224XZ37 A0A034V031 A0A0L7RHX1 A0A0K8V7H3 A0A158NB79 A0A0A1XE51 A0A1A9YIH3 N6T8S3 A0A1I8Q9G9 W8BLB7 A0A1B0AKS2 U4UJF9 A0A067R6E8 A0A1A9WLK0 F4X698 A0A1A9UG58 A0A151IDY9 A0A1A9ZKD4 A0A1B0FDP0 B4HU10 A0A0A9VWZ3 A0A151X314 A0A1I8MPY4 A0A212F547 A0A182G1N0 B0WB60 Q17J66 T1GEJ5 A0A084W000 A0A182IWB1 W5JCQ1 A0A182KA83 A0A182FQS9 X1XHE5 A0A195FTP4 A0A182RNX5 A0A182XHQ6 A0A182W001 A0A182UMY6 A0A182LGH9 A0A182HTN2 A0A182LRX7 Q7QCV2 A0A182Y447 A0A182SKQ3 A0A1S3DSP4 A0A0M3QWE9 A0A151JCB9 A0A182TUV2 A0A2H8TX95 E9J5F6 E2BTC3 A0A336MLD1 A0A182Q6P9 I4DMM2 A0A182P272 E0VIF7 A0A336M1M4 A0A1W4W872 E1ZUU6 A0A182N776 A0A3B0JXJ3 A0A2J7R1K2 A0A3B0JPM7 B4LB91 Q9VZH2-2 Q9VZH2 B3NG45 B4QQG6 B4PHQ5 B4J1F0 B4L9D3 A0A1J1HYT9 A0A2W1BQR2 A0A0J9RP82 A0A3L8DLM6 Q29D77 B4N3K3 A0A026WTF4 B4H7V6 A0A146L7U5 A0A1V1G575 B3M4V9 A0A1S3DWF4 A0A2H8TJX9 A0A194Q2L6 T1HZE5 A0A2A4K3W1 A0A2S2R3Z0 A0A1W4X873 A0A2P8Z9E4
PDB
5GVT
E-value=6.91052e-19,
Score=224
Ontologies
GO
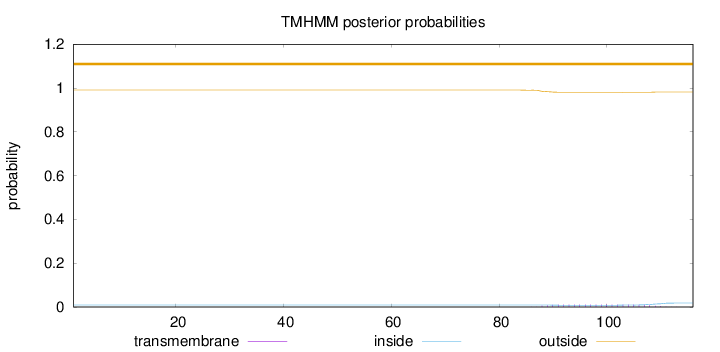
Topology
Subcellular location
Secreted
Localizes to vesicle-like structures in epidermal cells. With evidence from 11 publications.
Cell projection Localizes to vesicle-like structures in epidermal cells. With evidence from 11 publications.
Axon Localizes to vesicle-like structures in epidermal cells. With evidence from 11 publications.
Cell projection Localizes to vesicle-like structures in epidermal cells. With evidence from 11 publications.
Axon Localizes to vesicle-like structures in epidermal cells. With evidence from 11 publications.
Length:
116
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22705
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00939
outside
1 - 116
Population Genetic Test Statistics
Pi
142.564487
Theta
126.604935
Tajima's D
-0.501236
CLR
0
CSRT
0.246687665616719
Interpretation
Uncertain
