Gene
KWMTBOMO04175
Pre Gene Modal
BGIBMGA008726
Annotation
PREDICTED:_kinesin-like_protein_KLP2_[Bombyx_mori]
Full name
Kinesin-like protein
+ More
Kinesin-like protein Klp61F
Kinesin-like protein Klp61F
Alternative Name
Bipolar kinesin KRP-130
Location in the cell
Nuclear Reliability : 2.508
Sequence
CDS
ATGACGTCGGAGAAAGGATGTCGGAAAGAAAAGAATCAAAATATCCAAGTGTTTGTTCGTTTGAGACCGCTCAACCAACGGGAACGAGACCTCAAATCTTTGGGGGTAGTTGAAGTGCACAATAACAAGGAGGTTGTTGTGAGGATCTCACAGCAGAACAGCATTACCAAGAAGTTCACCTTTGACAGAGCATTCGCTCCTTATGCTAACCAGGTGGAAGTTTACCAAGAGGTGGTCAGCCCACTCATTGAAGAAGTGCTGGCTGGCTACAATTGCACTGTCTTTGCTTACGGTCAGACAGGCACTGGCAAGACCCACACAATGGTCGGAGAAAATACTGGTGATGAAACTACTTGGCAAAAGGATCCATTAGCCGGTATAATACCACGTGCGCTGAGCCAACTTTTTGATGAGCTTCGTATTTCTAACACCGAGTACACGGTCCGTGTGTCTTACTTGGAACTATATAATGAAGAGCTCTTTGACTTGCTGGCAACATCTGAAGACAATTCCAAGCTCAGGATTTATGAAGATGTCACTAGGAAAGGATCTAATATTGTAAATGGGCTAGAAGAGATAACTGTCTACAACAAAAAGGAAGTGTTCAGAATTATGGCCCAAGGACAGGAAAGGAAGAAAGTGGCTTCGACACTCATGAACGCACAGTCCAGTCGCTCCCACACCGTCTTCACGATCGTAGTCCACATGAAAGAAAACAGTCCTGAGGGCGAGGAGCTGGTGAAGATAGGCAAGCTAAACCTGGTGGATCTCGCTGGCTCAGAGAACATCAGCAAAGCTGGTTCTGACAACCCAGCCAAGCGGGAGAGGGCCCGGGAGTGTGTGAACATCAATCAGTCTCTGCTAACTCTTGGGAGGGTCATCACTGCACTTGTGGAAAGGCATCCTCATGTTCCTTATAGAGAATCAAAACTAACCAGGATCCTGCAAGAATCCCTCGGTGGTAGAACCAAAACTTCAATTATTGCTACAATCTCACCTGGACATAAGGACCTGGAAGAGACCATGAGCACTCTTGAGTATGCTCACAGAGCAAAGAACATTCAGAACAAGCCTGAGATCAACCAGAAAATGACAAAGAAAGCTATATTGAAAGAATATGCTGAAGAAATTGACAGATTGAAGAGGGATCTGCAAGCTGCAAGGGATAAAAATGGAGTATATCTAGCGAATGAAACCTTCGCCGAGATAACATTGAAACAGGAAGAACAAAGAAAAGAAATACAAGAACTCTTACTAAAGAAGAGAGCGATGGAAGAGGAAAGAGATAAGATGGAGAATGTCTTTAGTAACTTGAGTCGACAGTTGGAGTGCAGGGATGCCGAGCTCAATGAAACAAAGAATAAATTGAACAAAACAAATGCGAGATTAGCAGACACCTGTAATGTTCTGAAGACAACAAAACAACAATTTGAAGAGCAACAGTATTTAGTTCTGAAACATCGTGATACAGAACAAACTCTCACTCAACAAGCGAATCAGTTGCTGGCCACAGTAGACGTCGCCACCACACATGTCGCTTGCTTGCATGATAGTGTTGAAAAGAGGAGGCAATTAGAAAATGTTAATTTAGAAGTGACTAAAACTTACGTTGAACAATCCTCACAACACCGGGAGCACCTCACCGCTGGCATTGGGCATTACGTAGAGAACATCACTGCGGCACTGAAATCCCTCGATGATGCTATGAATGACTATACAGACAATAACACTCGCACACAAGAAGAAAGCAAACTTAAATTGGAAGAGTTAATTGGAATGATGGAGACAACTATCAATGAGATAAGATCCCTAGCATTGGACACACAGACCTCATTGGTTACAACAACAGAACAACATCGAAACGAAGCTCGGTCGATGGTAAGCGACCACAAGATGGCAATCTGTGCTGTTGGAGATTCATTAGTGGAGAGTGCTAACGATACCTTAGTCGCGGCCACCAACGTGGACCTCAGAGGACACATCGCTAGATTTGTCGAGCAGTGGTACAGGGGAGCGGCGGAGCGCGTGACGTCACTGCAGCGTGCGCACGCAGCGTTCGACACGCGCGGGCGGGACGCGCGGCTGGCACGCGGCGTGCTGCACGCGGCCCGGCGGGACGGCGCGCGGGAGCAGGCGGCGCGCGCGCGTCTCGCGGTGCAAGCGTGTCTACAGGTAGTCAATCAAATCAAATGA
Protein
MTSEKGCRKEKNQNIQVFVRLRPLNQRERDLKSLGVVEVHNNKEVVVRISQQNSITKKFTFDRAFAPYANQVEVYQEVVSPLIEEVLAGYNCTVFAYGQTGTGKTHTMVGENTGDETTWQKDPLAGIIPRALSQLFDELRISNTEYTVRVSYLELYNEELFDLLATSEDNSKLRIYEDVTRKGSNIVNGLEEITVYNKKEVFRIMAQGQERKKVASTLMNAQSSRSHTVFTIVVHMKENSPEGEELVKIGKLNLVDLAGSENISKAGSDNPAKRERARECVNINQSLLTLGRVITALVERHPHVPYRESKLTRILQESLGGRTKTSIIATISPGHKDLEETMSTLEYAHRAKNIQNKPEINQKMTKKAILKEYAEEIDRLKRDLQAARDKNGVYLANETFAEITLKQEEQRKEIQELLLKKRAMEEERDKMENVFSNLSRQLECRDAELNETKNKLNKTNARLADTCNVLKTTKQQFEEQQYLVLKHRDTEQTLTQQANQLLATVDVATTHVACLHDSVEKRRQLENVNLEVTKTYVEQSSQHREHLTAGIGHYVENITAALKSLDDAMNDYTDNNTRTQEESKLKLEELIGMMETTINEIRSLALDTQTSLVTTTEQHRNEARSMVSDHKMAICAVGDSLVESANDTLVAATNVDLRGHIARFVEQWYRGAAERVTSLQRAHAAFDTRGRDARLARGVLHAARRDGAREQAARARLAVQACLQVVNQIK
Summary
Description
Important role in mitotic dividing cells (PubMed:8227131). Microtubule motor required for spindle body separation (PubMed:8918872). Slow plus-end directed microtubule motor capable of cross-linking and sliding apart antiparallel microtubules, thereby pushing apart the associated spindle poles during spindle assembly and function (PubMed:8918872, PubMed:8589456, PubMed:19062285). Forms cross-links between microtubules within interpolar microtubule bundles (PubMed:9885249, PubMed:19158379). Contributes to the length of the metaphase spindle, maintains the prometaphase spindle by antagonizing Ncd, drives anaphase B, and also contributes to normal chromosome congression, kinetochore spacing, and anaphase A rates (PubMed:19158379). Displays microtubule-stimulated ATPase activity (PubMed:8589456). Required for normal fusome organization (PubMed:10469596). Required in non-mitotic cells for transport of secretory proteins from the Golgi complex to the cell surface (PubMed:23857769).
Subunit
Homotetramer (PubMed:8538794, PubMed:9885249, PubMed:24714498). Consists of two pairs of polypeptides associated by coiled-coil interactions to form two homodimers (PubMed:8538794). The homodimers are linked by lateral interactions between their coiled-coil regions to form a bipolar homotetramer consisting of a central rod with two motor domains projecting from either end (PubMed:8538794). Parallel coiled coils extend from each pair of motor heads, switch to two antiparallel coiled coils in the central region and then back to parallel coiled coils (PubMed:24714498). Interacts with Wee1 (PubMed:19800237).
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. BimC subfamily.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. BimC subfamily.
Keywords
3D-structure
ATP-binding
Cell cycle
Cell division
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Direct protein sequencing
Microtubule
Mitosis
Motor protein
Nucleotide-binding
Phosphoprotein
Reference proteome
Feature
chain Kinesin-like protein Klp61F
Uniprot
H9JGS9
A0A2A4K3W6
A0A3S2NYV7
A0A2H1WJI6
A0A212EIG2
A0A194PW97
+ More
A0A194RCC7 A0A2W1BXA5 A0A1I8P378 P46863 Q0PQ30 B4HVQ1 A0A0J9RLX7 B4QLY4 B3NB76 Q0PQ31 B3M537 B4LG86 B4PD55 B4J2F6 A0A3B0JRK3 Q29EV0 A0A1W4W768 B4L0M5 A0A0L0CR22 A0A1B0A2F7 A0A1I8NCE9 A0A1A9W4K9 A0A0A1WPI6 W8BFY4 A0A0M4EJL6 A0A0K8TXE6 A0A336LMR6 A0A2J7QSN9 A0A1B0BAB5 A0A182KRE0 A0A182TUA3 A0A182IP87 A0A182I3C6 A0A182V4Z5 Q7QJG2 A0A1B0C8R8 A0A182X3P3 A0A182QBE6 A0A182XYY0 A0A182FF61 A0A232FJ27 K7J300 A0A182PQD4 A0A182KAR0 B4MXY8 A0A182MQK1 A0A182RSQ1 A0A084WNC6 A0A195F4N0 A0A158NE35 A0A182WJ67 A0A0P5C224 A0A0P5C278 A0A0N8A9Q9 A0A0P5EK76 A0A0P6GJC8 A0A0P6FH74 A0A0P5H1R0 A0A0P5WQL3 A0A0P4YVL0 A0A0P5PB87 A0A0P5GPT4 A0A0P5AHG7 A0A182NHT7 A0A195BP88 A0A0P5GVE6 A0A0P4WWK6 A0A0N8BKA1 A0A0P5F0I5 A0A0P4Z4S7 A0A0P4Z622 A0A0P5IL51 A0A0P6IHI9 A0A0N8CZQ2 A0A0N8CLC8 A0A0P5TY16 A0A0P5L4P1 A0A0P5EV44 A0A0P6CX10 A0A0N8EBY4 F4WZ40 A0A0P5WKJ5 A0A0P5R5R9 E9IYU5 A0A0P5XN86 E9GVD2 A0A182GX94 A0A0P4YC33 A0A0P5WWF5 Q16HA8
A0A194RCC7 A0A2W1BXA5 A0A1I8P378 P46863 Q0PQ30 B4HVQ1 A0A0J9RLX7 B4QLY4 B3NB76 Q0PQ31 B3M537 B4LG86 B4PD55 B4J2F6 A0A3B0JRK3 Q29EV0 A0A1W4W768 B4L0M5 A0A0L0CR22 A0A1B0A2F7 A0A1I8NCE9 A0A1A9W4K9 A0A0A1WPI6 W8BFY4 A0A0M4EJL6 A0A0K8TXE6 A0A336LMR6 A0A2J7QSN9 A0A1B0BAB5 A0A182KRE0 A0A182TUA3 A0A182IP87 A0A182I3C6 A0A182V4Z5 Q7QJG2 A0A1B0C8R8 A0A182X3P3 A0A182QBE6 A0A182XYY0 A0A182FF61 A0A232FJ27 K7J300 A0A182PQD4 A0A182KAR0 B4MXY8 A0A182MQK1 A0A182RSQ1 A0A084WNC6 A0A195F4N0 A0A158NE35 A0A182WJ67 A0A0P5C224 A0A0P5C278 A0A0N8A9Q9 A0A0P5EK76 A0A0P6GJC8 A0A0P6FH74 A0A0P5H1R0 A0A0P5WQL3 A0A0P4YVL0 A0A0P5PB87 A0A0P5GPT4 A0A0P5AHG7 A0A182NHT7 A0A195BP88 A0A0P5GVE6 A0A0P4WWK6 A0A0N8BKA1 A0A0P5F0I5 A0A0P4Z4S7 A0A0P4Z622 A0A0P5IL51 A0A0P6IHI9 A0A0N8CZQ2 A0A0N8CLC8 A0A0P5TY16 A0A0P5L4P1 A0A0P5EV44 A0A0P6CX10 A0A0N8EBY4 F4WZ40 A0A0P5WKJ5 A0A0P5R5R9 E9IYU5 A0A0P5XN86 E9GVD2 A0A182GX94 A0A0P4YC33 A0A0P5WWF5 Q16HA8
Pubmed
19121390
22118469
26354079
28756777
8227131
10731132
+ More
12537572 12537569 1924306 8918872 8589456 8538794 10469596 9885249 17372656 19062285 18327897 19800237 19158379 23857769 19285086 24714498 17994087 22936249 17550304 15632085 18057021 26108605 25315136 25830018 24495485 20966253 12364791 25244985 28648823 20075255 24438588 21347285 21719571 21282665 21292972 26483478 17510324
12537572 12537569 1924306 8918872 8589456 8538794 10469596 9885249 17372656 19062285 18327897 19800237 19158379 23857769 19285086 24714498 17994087 22936249 17550304 15632085 18057021 26108605 25315136 25830018 24495485 20966253 12364791 25244985 28648823 20075255 24438588 21347285 21719571 21282665 21292972 26483478 17510324
EMBL
BABH01034392
BABH01034393
BABH01034394
BABH01034395
BABH01034396
NWSH01000158
+ More
PCG78945.1 RSAL01000020 RVE52567.1 ODYU01009057 SOQ53198.1 AGBW02014657 OWR41268.1 KQ459589 KPI97651.1 KQ460367 KPJ15498.1 KZ149929 PZC77440.1 U01842 AE014296 AY069442 M74428 DQ782382 ABG91087.1 CH480817 EDW50016.1 CM002912 KMY96772.1 CM000363 EDX08770.1 CH954178 EDV50129.1 DQ782381 ABG91086.1 CH902618 EDV39516.1 CH940647 EDW69394.1 CM000159 EDW92803.1 CH916366 EDV96015.1 OUUW01000009 SPP84767.1 CH379070 EAL29960.1 CH933809 EDW18102.1 KRG05979.1 JRES01000036 KNC34696.1 GBXI01013535 JAD00757.1 GAMC01010697 GAMC01010693 JAB95858.1 CP012525 ALC44238.1 GDHF01033356 JAI18958.1 UFQT01000063 SSX19266.1 NEVH01011876 PNF31587.1 JXJN01010891 APCN01000205 AAAB01008807 EAA04655.4 AJWK01001311 AXCN02000377 NNAY01000174 OXU30307.1 CH963876 EDW76977.1 AXCM01000691 ATLV01024587 KE525352 KFB51720.1 KQ981820 KYN35132.1 ADTU01012730 GDIP01177228 JAJ46174.1 GDIP01177227 JAJ46175.1 GDIP01168665 JAJ54737.1 GDIP01145275 JAJ78127.1 GDIQ01033903 JAN60834.1 GDIQ01047845 JAN46892.1 GDIQ01233229 JAK18496.1 GDIP01083030 JAM20685.1 GDIP01232965 JAI90436.1 GDIQ01131026 JAL20700.1 GDIQ01244876 GDIQ01240941 GDIQ01220452 GDIQ01216987 GDIQ01140809 GDIQ01050761 JAK06849.1 GDIP01219520 GDIP01199467 GDIQ01187494 JAJ23935.1 JAK64231.1 KQ976424 KYM88325.1 GDIQ01242380 JAK09345.1 GDIP01254011 JAI69390.1 GDIQ01169451 JAK82274.1 GDIQ01267544 JAJ84180.1 GDIP01217964 GDIQ01032596 JAJ05438.1 JAN62141.1 GDIP01221773 JAJ01629.1 GDIQ01211844 JAK39881.1 GDIQ01010957 JAN83780.1 GDIP01083031 JAM20684.1 GDIP01120423 JAL83291.1 GDIP01120377 JAL83337.1 GDIQ01180449 JAK71276.1 GDIQ01267543 JAJ84181.1 GDIQ01086480 JAN08257.1 GDIQ01041796 JAN52941.1 GL888465 EGI60543.1 GDIP01084777 JAM18938.1 GDIQ01110622 JAL41104.1 GL767051 EFZ14248.1 GDIP01069628 JAM34087.1 GL732568 EFX76397.1 JXUM01094920 JXUM01094921 JXUM01094922 JXUM01094923 KQ564161 KXJ72656.1 GDIP01230567 JAI92834.1 GDIP01080981 JAM22734.1 CH478187 EAT33633.1
PCG78945.1 RSAL01000020 RVE52567.1 ODYU01009057 SOQ53198.1 AGBW02014657 OWR41268.1 KQ459589 KPI97651.1 KQ460367 KPJ15498.1 KZ149929 PZC77440.1 U01842 AE014296 AY069442 M74428 DQ782382 ABG91087.1 CH480817 EDW50016.1 CM002912 KMY96772.1 CM000363 EDX08770.1 CH954178 EDV50129.1 DQ782381 ABG91086.1 CH902618 EDV39516.1 CH940647 EDW69394.1 CM000159 EDW92803.1 CH916366 EDV96015.1 OUUW01000009 SPP84767.1 CH379070 EAL29960.1 CH933809 EDW18102.1 KRG05979.1 JRES01000036 KNC34696.1 GBXI01013535 JAD00757.1 GAMC01010697 GAMC01010693 JAB95858.1 CP012525 ALC44238.1 GDHF01033356 JAI18958.1 UFQT01000063 SSX19266.1 NEVH01011876 PNF31587.1 JXJN01010891 APCN01000205 AAAB01008807 EAA04655.4 AJWK01001311 AXCN02000377 NNAY01000174 OXU30307.1 CH963876 EDW76977.1 AXCM01000691 ATLV01024587 KE525352 KFB51720.1 KQ981820 KYN35132.1 ADTU01012730 GDIP01177228 JAJ46174.1 GDIP01177227 JAJ46175.1 GDIP01168665 JAJ54737.1 GDIP01145275 JAJ78127.1 GDIQ01033903 JAN60834.1 GDIQ01047845 JAN46892.1 GDIQ01233229 JAK18496.1 GDIP01083030 JAM20685.1 GDIP01232965 JAI90436.1 GDIQ01131026 JAL20700.1 GDIQ01244876 GDIQ01240941 GDIQ01220452 GDIQ01216987 GDIQ01140809 GDIQ01050761 JAK06849.1 GDIP01219520 GDIP01199467 GDIQ01187494 JAJ23935.1 JAK64231.1 KQ976424 KYM88325.1 GDIQ01242380 JAK09345.1 GDIP01254011 JAI69390.1 GDIQ01169451 JAK82274.1 GDIQ01267544 JAJ84180.1 GDIP01217964 GDIQ01032596 JAJ05438.1 JAN62141.1 GDIP01221773 JAJ01629.1 GDIQ01211844 JAK39881.1 GDIQ01010957 JAN83780.1 GDIP01083031 JAM20684.1 GDIP01120423 JAL83291.1 GDIP01120377 JAL83337.1 GDIQ01180449 JAK71276.1 GDIQ01267543 JAJ84181.1 GDIQ01086480 JAN08257.1 GDIQ01041796 JAN52941.1 GL888465 EGI60543.1 GDIP01084777 JAM18938.1 GDIQ01110622 JAL41104.1 GL767051 EFZ14248.1 GDIP01069628 JAM34087.1 GL732568 EFX76397.1 JXUM01094920 JXUM01094921 JXUM01094922 JXUM01094923 KQ564161 KXJ72656.1 GDIP01230567 JAI92834.1 GDIP01080981 JAM22734.1 CH478187 EAT33633.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000095300 UP000000803 UP000001292 UP000000304 UP000008711 UP000007801 UP000008792 UP000002282 UP000001070 UP000268350 UP000001819 UP000192221 UP000009192 UP000037069 UP000092445 UP000095301 UP000091820 UP000092553 UP000235965 UP000092460 UP000075882 UP000075902 UP000075880 UP000075840 UP000075903 UP000007062 UP000092461 UP000076407 UP000075886 UP000076408 UP000069272 UP000215335 UP000002358 UP000075885 UP000075881 UP000007798 UP000075883 UP000075900 UP000030765 UP000078541 UP000005205 UP000075920 UP000075884 UP000078540 UP000007755 UP000000305 UP000069940 UP000249989 UP000008820
UP000095300 UP000000803 UP000001292 UP000000304 UP000008711 UP000007801 UP000008792 UP000002282 UP000001070 UP000268350 UP000001819 UP000192221 UP000009192 UP000037069 UP000092445 UP000095301 UP000091820 UP000092553 UP000235965 UP000092460 UP000075882 UP000075902 UP000075880 UP000075840 UP000075903 UP000007062 UP000092461 UP000076407 UP000075886 UP000076408 UP000069272 UP000215335 UP000002358 UP000075885 UP000075881 UP000007798 UP000075883 UP000075900 UP000030765 UP000078541 UP000005205 UP000075920 UP000075884 UP000078540 UP000007755 UP000000305 UP000069940 UP000249989 UP000008820
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9JGS9
A0A2A4K3W6
A0A3S2NYV7
A0A2H1WJI6
A0A212EIG2
A0A194PW97
+ More
A0A194RCC7 A0A2W1BXA5 A0A1I8P378 P46863 Q0PQ30 B4HVQ1 A0A0J9RLX7 B4QLY4 B3NB76 Q0PQ31 B3M537 B4LG86 B4PD55 B4J2F6 A0A3B0JRK3 Q29EV0 A0A1W4W768 B4L0M5 A0A0L0CR22 A0A1B0A2F7 A0A1I8NCE9 A0A1A9W4K9 A0A0A1WPI6 W8BFY4 A0A0M4EJL6 A0A0K8TXE6 A0A336LMR6 A0A2J7QSN9 A0A1B0BAB5 A0A182KRE0 A0A182TUA3 A0A182IP87 A0A182I3C6 A0A182V4Z5 Q7QJG2 A0A1B0C8R8 A0A182X3P3 A0A182QBE6 A0A182XYY0 A0A182FF61 A0A232FJ27 K7J300 A0A182PQD4 A0A182KAR0 B4MXY8 A0A182MQK1 A0A182RSQ1 A0A084WNC6 A0A195F4N0 A0A158NE35 A0A182WJ67 A0A0P5C224 A0A0P5C278 A0A0N8A9Q9 A0A0P5EK76 A0A0P6GJC8 A0A0P6FH74 A0A0P5H1R0 A0A0P5WQL3 A0A0P4YVL0 A0A0P5PB87 A0A0P5GPT4 A0A0P5AHG7 A0A182NHT7 A0A195BP88 A0A0P5GVE6 A0A0P4WWK6 A0A0N8BKA1 A0A0P5F0I5 A0A0P4Z4S7 A0A0P4Z622 A0A0P5IL51 A0A0P6IHI9 A0A0N8CZQ2 A0A0N8CLC8 A0A0P5TY16 A0A0P5L4P1 A0A0P5EV44 A0A0P6CX10 A0A0N8EBY4 F4WZ40 A0A0P5WKJ5 A0A0P5R5R9 E9IYU5 A0A0P5XN86 E9GVD2 A0A182GX94 A0A0P4YC33 A0A0P5WWF5 Q16HA8
A0A194RCC7 A0A2W1BXA5 A0A1I8P378 P46863 Q0PQ30 B4HVQ1 A0A0J9RLX7 B4QLY4 B3NB76 Q0PQ31 B3M537 B4LG86 B4PD55 B4J2F6 A0A3B0JRK3 Q29EV0 A0A1W4W768 B4L0M5 A0A0L0CR22 A0A1B0A2F7 A0A1I8NCE9 A0A1A9W4K9 A0A0A1WPI6 W8BFY4 A0A0M4EJL6 A0A0K8TXE6 A0A336LMR6 A0A2J7QSN9 A0A1B0BAB5 A0A182KRE0 A0A182TUA3 A0A182IP87 A0A182I3C6 A0A182V4Z5 Q7QJG2 A0A1B0C8R8 A0A182X3P3 A0A182QBE6 A0A182XYY0 A0A182FF61 A0A232FJ27 K7J300 A0A182PQD4 A0A182KAR0 B4MXY8 A0A182MQK1 A0A182RSQ1 A0A084WNC6 A0A195F4N0 A0A158NE35 A0A182WJ67 A0A0P5C224 A0A0P5C278 A0A0N8A9Q9 A0A0P5EK76 A0A0P6GJC8 A0A0P6FH74 A0A0P5H1R0 A0A0P5WQL3 A0A0P4YVL0 A0A0P5PB87 A0A0P5GPT4 A0A0P5AHG7 A0A182NHT7 A0A195BP88 A0A0P5GVE6 A0A0P4WWK6 A0A0N8BKA1 A0A0P5F0I5 A0A0P4Z4S7 A0A0P4Z622 A0A0P5IL51 A0A0P6IHI9 A0A0N8CZQ2 A0A0N8CLC8 A0A0P5TY16 A0A0P5L4P1 A0A0P5EV44 A0A0P6CX10 A0A0N8EBY4 F4WZ40 A0A0P5WKJ5 A0A0P5R5R9 E9IYU5 A0A0P5XN86 E9GVD2 A0A182GX94 A0A0P4YC33 A0A0P5WWF5 Q16HA8
PDB
2WBE
E-value=1.22695e-129,
Score=1189
Ontologies
KEGG
GO
GO:0007018
GO:0008017
GO:0003777
GO:0005524
GO:0003774
GO:0005874
GO:1990498
GO:0005875
GO:0005783
GO:0005818
GO:0051301
GO:0097431
GO:0008574
GO:0000278
GO:0045478
GO:0072686
GO:0051289
GO:0007052
GO:0051299
GO:0005794
GO:0045169
GO:0005634
GO:0005737
GO:0007100
GO:0001578
GO:0042998
GO:0005819
GO:0009306
GO:0005871
GO:0031535
GO:0007030
GO:0090307
GO:0016887
GO:0005876
GO:0016021
GO:0042555
GO:0006260
GO:0004672
GO:0048384
PANTHER
Topology
Subcellular location
Cytoplasm
In somatic cells, cytoplasmic during interphase, localized to centrosomal asters at the onset of mitosis in prophase and associated with spindle structures during the remainder of mitosis (PubMed:10469596). In male and female germ cells, associates with fusomes during interphase, then localizes to centrosomal asters during prophase and to spindles in metaphase (PubMed:10469596). Coincident with spindle microtubules from prophase to metaphase and, as mitosis proceeds, found both at the spindle poles and along the spindle fibers (PubMed:8589456). With evidence from 5 publications.
Cytoskeleton In somatic cells, cytoplasmic during interphase, localized to centrosomal asters at the onset of mitosis in prophase and associated with spindle structures during the remainder of mitosis (PubMed:10469596). In male and female germ cells, associates with fusomes during interphase, then localizes to centrosomal asters during prophase and to spindles in metaphase (PubMed:10469596). Coincident with spindle microtubules from prophase to metaphase and, as mitosis proceeds, found both at the spindle poles and along the spindle fibers (PubMed:8589456). With evidence from 5 publications.
Spindle In somatic cells, cytoplasmic during interphase, localized to centrosomal asters at the onset of mitosis in prophase and associated with spindle structures during the remainder of mitosis (PubMed:10469596). In male and female germ cells, associates with fusomes during interphase, then localizes to centrosomal asters during prophase and to spindles in metaphase (PubMed:10469596). Coincident with spindle microtubules from prophase to metaphase and, as mitosis proceeds, found both at the spindle poles and along the spindle fibers (PubMed:8589456). With evidence from 5 publications.
Spindle pole In somatic cells, cytoplasmic during interphase, localized to centrosomal asters at the onset of mitosis in prophase and associated with spindle structures during the remainder of mitosis (PubMed:10469596). In male and female germ cells, associates with fusomes during interphase, then localizes to centrosomal asters during prophase and to spindles in metaphase (PubMed:10469596). Coincident with spindle microtubules from prophase to metaphase and, as mitosis proceeds, found both at the spindle poles and along the spindle fibers (PubMed:8589456). With evidence from 5 publications.
Cytoskeleton In somatic cells, cytoplasmic during interphase, localized to centrosomal asters at the onset of mitosis in prophase and associated with spindle structures during the remainder of mitosis (PubMed:10469596). In male and female germ cells, associates with fusomes during interphase, then localizes to centrosomal asters during prophase and to spindles in metaphase (PubMed:10469596). Coincident with spindle microtubules from prophase to metaphase and, as mitosis proceeds, found both at the spindle poles and along the spindle fibers (PubMed:8589456). With evidence from 5 publications.
Spindle In somatic cells, cytoplasmic during interphase, localized to centrosomal asters at the onset of mitosis in prophase and associated with spindle structures during the remainder of mitosis (PubMed:10469596). In male and female germ cells, associates with fusomes during interphase, then localizes to centrosomal asters during prophase and to spindles in metaphase (PubMed:10469596). Coincident with spindle microtubules from prophase to metaphase and, as mitosis proceeds, found both at the spindle poles and along the spindle fibers (PubMed:8589456). With evidence from 5 publications.
Spindle pole In somatic cells, cytoplasmic during interphase, localized to centrosomal asters at the onset of mitosis in prophase and associated with spindle structures during the remainder of mitosis (PubMed:10469596). In male and female germ cells, associates with fusomes during interphase, then localizes to centrosomal asters during prophase and to spindles in metaphase (PubMed:10469596). Coincident with spindle microtubules from prophase to metaphase and, as mitosis proceeds, found both at the spindle poles and along the spindle fibers (PubMed:8589456). With evidence from 5 publications.
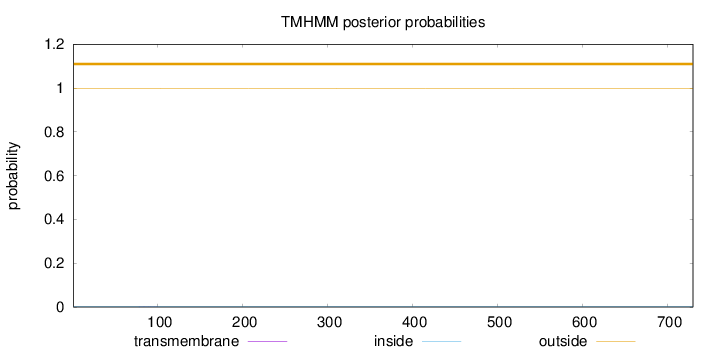
Length:
730
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01034
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00111
outside
1 - 730
Population Genetic Test Statistics
Pi
26.792513
Theta
25.667299
Tajima's D
1.300669
CLR
0.360958
CSRT
0.744262786860657
Interpretation
Uncertain
