Gene
KWMTBOMO04172 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008668
Annotation
clip_domain_serine_protease_4_precursor_[Bombyx_mori]
Full name
Venom serine protease Bi-VSP
+ More
Venom protease
Venom protease
Alternative Name
Thrombin-like enzyme
Location in the cell
Cytoplasmic Reliability : 1.804 Extracellular Reliability : 1.502
Sequence
CDS
ATGTTGAATACGTATTTCACTCAAAATAGTACTCGTATTTGCCACGGCTACACCAAATTTCACGAAGACTTCCGAAGAAATTATCGCGAATCCTTAATTTCCAGGTACTTAGTGCGAGTGGGAGAGCTGGATCTTGCCAGAGACGATGAAGGTGCTACTCCTGTGGACGTTCTCATTAAGACGAAGATCAAGCACGAGCAATACGATGCCGCCTCTTATACCAACGACATCGGCATCCTGGTGCTGGAGAAGGACGTACCTATTACAGACTTGATTAAGCCTATCTGCATCCCCAAGGACACAGAGCTTCGCTCCCGCAGCTTCGAGGACTACAACCCCATCATAGCCGGCTGGGGGGACACAGAGTTCCGCGGTCCATCCGCAACGCACCTCCAAGTCCTGCAGCTGCCTGTGGTCAGCAACGACTTTTGTGCTCAAGCGTACTCGCCATATAAGAATCAGAAGATCGATGAGAGGGTGCTCTGCGCTGGGTACAAGAAGGGGGGCAAGGACGCCTGTCAGGGGGACAGCGGGGGCCCTCTGATGCAGCCCATTTGGAACTCGCAGACCTACAAGACCTACTTCTACCAGATCGGAGTGGTCTCCTTCGGCAAGAAATGCGCTGAGGCTGGATTCCCCGGCGTCTATGCCAGGGTCACGCACTTCGTTCCCTGGATCCAAGAGAAGGTTGTGGGTCATGCGTGA
Protein
MLNTYFTQNSTRICHGYTKFHEDFRRNYRESLISRYLVRVGELDLARDDEGATPVDVLIKTKIKHEQYDAASYTNDIGILVLEKDVPITDLIKPICIPKDTELRSRSFEDYNPIIAGWGDTEFRGPSATHLQVLQLPVVSNDFCAQAYSPYKNQKIDERVLCAGYKKGGKDACQGDSGGPLMQPIWNSQTYKTYFYQIGVVSFGKKCAEAGFPGVYARVTHFVPWIQEKVVGHA
Summary
Description
Multifunctional venom serine protease. In insects, it acts as an arthropod prophenoloxidase-activating factor, thereby triggering the phenoloxidase cascade. When injected into larvae, it induces a lethal melanization response in target insects by modulating the innate immune response. In mammals, it converts fibrinogen into fibrin, activates prothrombin, and also degrades fibrin. In mammal, it may act in a cooperative manner with the serine protease inhibitor Bi-KTI (AC G3LH89) to promote the spread of bee venom under anti-bleeding conditions.
Miscellaneous
Does not activate plasminogen.
Similarity
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family. CLIP subfamily.
Belongs to the peptidase S1 family. CLIP subfamily.
Keywords
Blood coagulation cascade activating toxin
Calcium
Direct protein sequencing
Disulfide bond
Fibrinolytic toxin
Glycoprotein
Hemostasis impairing toxin
Hydrolase
Metal-binding
Protease
Prothrombin activator
Secreted
Serine protease
Signal
Toxin
Zymogen
Allergen
Feature
chain Venom serine protease Bi-VSP
Uniprot
Q8MP08
H9JGM1
A0A2A4JS27
A0A2W1BQQ9
Q8I927
Q5MPB7
+ More
Q5MPB8 Q8I925 Q8I926 A0A212F528 A0A2H1WWB5 A0A194PXA8 A0A212EIG1 A0A212EPS0 A0A212EIF7 A0A2A3ENM0 A0A1I9UVU8 A0A212EPV1 A0A154PEP1 A0A088ADM5 A0A2J7R078 A0A2J7R073 A0A0M8ZP71 A0A1V1FYF4 A0A1W4WNW1 A0A1W4WNW0 B0WB63 A0A1Y1M8P7 A0A067RFG3 K7J1M7 E2BNP2 A0A182GQG7 Q17J63 A0A0L7RGN5 E9J1L2 A0A1S4F0R9 A0A232EUU8 A0A0P4VP51 A0A1B6EF69 T1HGB7 A0A226EEU3 A0A026VUK8 W5JGA9 U4TZ65 A0A224XE75 A0A0P4ZFV9 A0A158P3U7 B5U2W0 F4WKX6 F1CGQ2 F1CGQ3 G3GQP4 A0A0P5H1N4 A0A0P5J354 A0A0P6DSV9 A0A0P6F026 A0A0P5LAI0 A0A164XDX4 A0A0P6CM36 A0A0P6GSZ8 A0A0P5QER7 A0A0P5YT07 K7J1M8 A0A0P5S303 A0A151WSE3 A0A0P5PRB7 A0A084W007 R4FQA1 B8QQQ1 A0A1S4EMH0 F5HKL0 E9H0K2 A0A182SD58 A0A067XL24 A0A195CNZ7 A0A0L7LIA9 A0A195DZM2 A0A195ATJ4 A0A2H8TVY8 D6W808 A0A151K0E7 A0A182LGK1 A0A182UXQ5 A0A291C3B4 K7J0K5 A0A182Q6I6 J9JVY2 Q7M4I3 A0A1D2MGY3 A0A232F2M8 A0A348G5Y4 A0A0P6G9Q0 A0A1W6EW33 A0A2R7VPE0 A0A0J7KRY6 A0A1B6DCP0 A0A1B6E0S4 A0A2P2I6P9 A0A0P6I905
Q5MPB8 Q8I925 Q8I926 A0A212F528 A0A2H1WWB5 A0A194PXA8 A0A212EIG1 A0A212EPS0 A0A212EIF7 A0A2A3ENM0 A0A1I9UVU8 A0A212EPV1 A0A154PEP1 A0A088ADM5 A0A2J7R078 A0A2J7R073 A0A0M8ZP71 A0A1V1FYF4 A0A1W4WNW1 A0A1W4WNW0 B0WB63 A0A1Y1M8P7 A0A067RFG3 K7J1M7 E2BNP2 A0A182GQG7 Q17J63 A0A0L7RGN5 E9J1L2 A0A1S4F0R9 A0A232EUU8 A0A0P4VP51 A0A1B6EF69 T1HGB7 A0A226EEU3 A0A026VUK8 W5JGA9 U4TZ65 A0A224XE75 A0A0P4ZFV9 A0A158P3U7 B5U2W0 F4WKX6 F1CGQ2 F1CGQ3 G3GQP4 A0A0P5H1N4 A0A0P5J354 A0A0P6DSV9 A0A0P6F026 A0A0P5LAI0 A0A164XDX4 A0A0P6CM36 A0A0P6GSZ8 A0A0P5QER7 A0A0P5YT07 K7J1M8 A0A0P5S303 A0A151WSE3 A0A0P5PRB7 A0A084W007 R4FQA1 B8QQQ1 A0A1S4EMH0 F5HKL0 E9H0K2 A0A182SD58 A0A067XL24 A0A195CNZ7 A0A0L7LIA9 A0A195DZM2 A0A195ATJ4 A0A2H8TVY8 D6W808 A0A151K0E7 A0A182LGK1 A0A182UXQ5 A0A291C3B4 K7J0K5 A0A182Q6I6 J9JVY2 Q7M4I3 A0A1D2MGY3 A0A232F2M8 A0A348G5Y4 A0A0P6G9Q0 A0A1W6EW33 A0A2R7VPE0 A0A0J7KRY6 A0A1B6DCP0 A0A1B6E0S4 A0A2P2I6P9 A0A0P6I905
EC Number
3.4.21.-
Pubmed
19121390
28756777
15944088
22118469
26354079
28410430
+ More
28004739 24845553 20075255 20798317 26483478 17510324 21282665 28648823 27129103 24508170 30249741 20920257 23761445 23537049 21347285 20454652 22359676 21719571 24438588 12364791 14747013 17210077 21292972 26227816 18362917 19820115 20966253 27289101
28004739 24845553 20075255 20798317 26483478 17510324 21282665 28648823 27129103 24508170 30249741 20920257 23761445 23537049 21347285 20454652 22359676 21719571 24438588 12364791 14747013 17210077 21292972 26227816 18362917 19820115 20966253 27289101
EMBL
AB073673
BAB91156.1
BABH01034383
BABH01034384
NWSH01000785
PCG74210.1
+ More
KZ149929 PZC77442.1 AY061933 AAL31704.1 AY672793 AAV91015.1 AY672792 AAV91014.1 AY061935 AAL31706.1 AY061934 AAL31705.1 AGBW02010265 OWR48842.1 ODYU01011542 SOQ57348.1 KQ459589 KPI97653.1 AGBW02014657 OWR41265.1 AGBW02013393 OWR43495.1 OWR41269.1 KZ288206 PBC33104.1 KU668683 APA05228.1 OWR43494.1 KQ434875 KZC09758.1 NEVH01008287 PNF34238.1 PNF34237.1 KQ435944 KOX68265.1 FX985286 BAX07299.1 DS231877 EDS42128.1 GEZM01037396 JAV82073.1 KK852669 KDR18888.1 GL449438 EFN82690.1 JXUM01080533 KQ563191 KXJ74306.1 CH477234 EAT46744.1 KQ414596 KOC69988.1 GL767674 EFZ13255.1 NNAY01002080 OXU22140.1 GDKW01003671 JAI52924.1 GEDC01000715 JAS36583.1 ACPB03015962 LNIX01000004 OXA55910.1 KK107847 QOIP01000005 EZA47437.1 RLU23132.1 ADMH02001605 ETN61854.1 KB631792 ERL86132.1 GFTR01005660 JAW10766.1 GDIP01213944 JAJ09458.1 ADTU01008561 FJ159442 FJ159443 GL888206 EGI65218.1 HQ257242 ADY75779.1 HQ257243 ADY75780.1 JF979158 JF979159 AEN41590.1 GDIQ01233256 JAK18469.1 GDIQ01211809 JAK39916.1 GDIQ01073307 JAN21430.1 GDIQ01076108 GDIQ01067715 JAN27022.1 GDIQ01172773 JAK78952.1 LRGB01001005 KZS14121.1 GDIQ01090555 JAN04182.1 GDIQ01211810 GDIQ01076107 GDIQ01057117 GDIQ01049775 GDIQ01030295 JAN64442.1 GDIQ01136492 JAL15234.1 GDIP01053817 JAM49898.1 GDIQ01093048 JAL58678.1 KQ982783 KYQ50711.1 GDIQ01125343 JAL26383.1 ATLV01019066 KE525259 KFB43551.1 GAHY01000519 JAA76991.1 EU665686 ACF70480.1 AAAB01008859 EGK96762.1 GL732581 EFX74738.1 JX399874 AGI44422.1 KQ977513 KYN02207.1 JTDY01001069 KOB74971.1 KQ980044 KYN18074.1 KQ976746 KYM75330.1 GFXV01005523 MBW17328.1 KQ971307 EFA11581.1 KQ981292 KYN43137.1 KX950713 ATF27964.1 AXCN02000328 ABLF02021787 LJIJ01001282 ODM92260.1 NNAY01001198 OXU24769.1 FX985522 BBF97857.1 GDIQ01036590 JAN58147.1 KY563523 ARK19932.1 KK854015 PTY09392.1 LBMM01003785 KMQ93137.1 GEDC01013824 JAS23474.1 GEDC01005772 JAS31526.1 IACF01004094 LAB69689.1 GDIQ01007827 JAN86910.1
KZ149929 PZC77442.1 AY061933 AAL31704.1 AY672793 AAV91015.1 AY672792 AAV91014.1 AY061935 AAL31706.1 AY061934 AAL31705.1 AGBW02010265 OWR48842.1 ODYU01011542 SOQ57348.1 KQ459589 KPI97653.1 AGBW02014657 OWR41265.1 AGBW02013393 OWR43495.1 OWR41269.1 KZ288206 PBC33104.1 KU668683 APA05228.1 OWR43494.1 KQ434875 KZC09758.1 NEVH01008287 PNF34238.1 PNF34237.1 KQ435944 KOX68265.1 FX985286 BAX07299.1 DS231877 EDS42128.1 GEZM01037396 JAV82073.1 KK852669 KDR18888.1 GL449438 EFN82690.1 JXUM01080533 KQ563191 KXJ74306.1 CH477234 EAT46744.1 KQ414596 KOC69988.1 GL767674 EFZ13255.1 NNAY01002080 OXU22140.1 GDKW01003671 JAI52924.1 GEDC01000715 JAS36583.1 ACPB03015962 LNIX01000004 OXA55910.1 KK107847 QOIP01000005 EZA47437.1 RLU23132.1 ADMH02001605 ETN61854.1 KB631792 ERL86132.1 GFTR01005660 JAW10766.1 GDIP01213944 JAJ09458.1 ADTU01008561 FJ159442 FJ159443 GL888206 EGI65218.1 HQ257242 ADY75779.1 HQ257243 ADY75780.1 JF979158 JF979159 AEN41590.1 GDIQ01233256 JAK18469.1 GDIQ01211809 JAK39916.1 GDIQ01073307 JAN21430.1 GDIQ01076108 GDIQ01067715 JAN27022.1 GDIQ01172773 JAK78952.1 LRGB01001005 KZS14121.1 GDIQ01090555 JAN04182.1 GDIQ01211810 GDIQ01076107 GDIQ01057117 GDIQ01049775 GDIQ01030295 JAN64442.1 GDIQ01136492 JAL15234.1 GDIP01053817 JAM49898.1 GDIQ01093048 JAL58678.1 KQ982783 KYQ50711.1 GDIQ01125343 JAL26383.1 ATLV01019066 KE525259 KFB43551.1 GAHY01000519 JAA76991.1 EU665686 ACF70480.1 AAAB01008859 EGK96762.1 GL732581 EFX74738.1 JX399874 AGI44422.1 KQ977513 KYN02207.1 JTDY01001069 KOB74971.1 KQ980044 KYN18074.1 KQ976746 KYM75330.1 GFXV01005523 MBW17328.1 KQ971307 EFA11581.1 KQ981292 KYN43137.1 KX950713 ATF27964.1 AXCN02000328 ABLF02021787 LJIJ01001282 ODM92260.1 NNAY01001198 OXU24769.1 FX985522 BBF97857.1 GDIQ01036590 JAN58147.1 KY563523 ARK19932.1 KK854015 PTY09392.1 LBMM01003785 KMQ93137.1 GEDC01013824 JAS23474.1 GEDC01005772 JAS31526.1 IACF01004094 LAB69689.1 GDIQ01007827 JAN86910.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000242457
UP000076502
+ More
UP000005203 UP000235965 UP000053105 UP000192223 UP000002320 UP000027135 UP000002358 UP000008237 UP000069940 UP000249989 UP000008820 UP000053825 UP000215335 UP000015103 UP000198287 UP000053097 UP000279307 UP000000673 UP000030742 UP000005205 UP000007755 UP000076858 UP000075809 UP000030765 UP000079169 UP000007062 UP000000305 UP000075901 UP000078542 UP000037510 UP000078492 UP000078540 UP000007266 UP000078541 UP000075882 UP000075903 UP000075886 UP000007819 UP000094527 UP000036403
UP000005203 UP000235965 UP000053105 UP000192223 UP000002320 UP000027135 UP000002358 UP000008237 UP000069940 UP000249989 UP000008820 UP000053825 UP000215335 UP000015103 UP000198287 UP000053097 UP000279307 UP000000673 UP000030742 UP000005205 UP000007755 UP000076858 UP000075809 UP000030765 UP000079169 UP000007062 UP000000305 UP000075901 UP000078542 UP000037510 UP000078492 UP000078540 UP000007266 UP000078541 UP000075882 UP000075903 UP000075886 UP000007819 UP000094527 UP000036403
Interpro
SUPFAM
SSF50494
SSF50494
Gene 3D
CDD
ProteinModelPortal
Q8MP08
H9JGM1
A0A2A4JS27
A0A2W1BQQ9
Q8I927
Q5MPB7
+ More
Q5MPB8 Q8I925 Q8I926 A0A212F528 A0A2H1WWB5 A0A194PXA8 A0A212EIG1 A0A212EPS0 A0A212EIF7 A0A2A3ENM0 A0A1I9UVU8 A0A212EPV1 A0A154PEP1 A0A088ADM5 A0A2J7R078 A0A2J7R073 A0A0M8ZP71 A0A1V1FYF4 A0A1W4WNW1 A0A1W4WNW0 B0WB63 A0A1Y1M8P7 A0A067RFG3 K7J1M7 E2BNP2 A0A182GQG7 Q17J63 A0A0L7RGN5 E9J1L2 A0A1S4F0R9 A0A232EUU8 A0A0P4VP51 A0A1B6EF69 T1HGB7 A0A226EEU3 A0A026VUK8 W5JGA9 U4TZ65 A0A224XE75 A0A0P4ZFV9 A0A158P3U7 B5U2W0 F4WKX6 F1CGQ2 F1CGQ3 G3GQP4 A0A0P5H1N4 A0A0P5J354 A0A0P6DSV9 A0A0P6F026 A0A0P5LAI0 A0A164XDX4 A0A0P6CM36 A0A0P6GSZ8 A0A0P5QER7 A0A0P5YT07 K7J1M8 A0A0P5S303 A0A151WSE3 A0A0P5PRB7 A0A084W007 R4FQA1 B8QQQ1 A0A1S4EMH0 F5HKL0 E9H0K2 A0A182SD58 A0A067XL24 A0A195CNZ7 A0A0L7LIA9 A0A195DZM2 A0A195ATJ4 A0A2H8TVY8 D6W808 A0A151K0E7 A0A182LGK1 A0A182UXQ5 A0A291C3B4 K7J0K5 A0A182Q6I6 J9JVY2 Q7M4I3 A0A1D2MGY3 A0A232F2M8 A0A348G5Y4 A0A0P6G9Q0 A0A1W6EW33 A0A2R7VPE0 A0A0J7KRY6 A0A1B6DCP0 A0A1B6E0S4 A0A2P2I6P9 A0A0P6I905
Q5MPB8 Q8I925 Q8I926 A0A212F528 A0A2H1WWB5 A0A194PXA8 A0A212EIG1 A0A212EPS0 A0A212EIF7 A0A2A3ENM0 A0A1I9UVU8 A0A212EPV1 A0A154PEP1 A0A088ADM5 A0A2J7R078 A0A2J7R073 A0A0M8ZP71 A0A1V1FYF4 A0A1W4WNW1 A0A1W4WNW0 B0WB63 A0A1Y1M8P7 A0A067RFG3 K7J1M7 E2BNP2 A0A182GQG7 Q17J63 A0A0L7RGN5 E9J1L2 A0A1S4F0R9 A0A232EUU8 A0A0P4VP51 A0A1B6EF69 T1HGB7 A0A226EEU3 A0A026VUK8 W5JGA9 U4TZ65 A0A224XE75 A0A0P4ZFV9 A0A158P3U7 B5U2W0 F4WKX6 F1CGQ2 F1CGQ3 G3GQP4 A0A0P5H1N4 A0A0P5J354 A0A0P6DSV9 A0A0P6F026 A0A0P5LAI0 A0A164XDX4 A0A0P6CM36 A0A0P6GSZ8 A0A0P5QER7 A0A0P5YT07 K7J1M8 A0A0P5S303 A0A151WSE3 A0A0P5PRB7 A0A084W007 R4FQA1 B8QQQ1 A0A1S4EMH0 F5HKL0 E9H0K2 A0A182SD58 A0A067XL24 A0A195CNZ7 A0A0L7LIA9 A0A195DZM2 A0A195ATJ4 A0A2H8TVY8 D6W808 A0A151K0E7 A0A182LGK1 A0A182UXQ5 A0A291C3B4 K7J0K5 A0A182Q6I6 J9JVY2 Q7M4I3 A0A1D2MGY3 A0A232F2M8 A0A348G5Y4 A0A0P6G9Q0 A0A1W6EW33 A0A2R7VPE0 A0A0J7KRY6 A0A1B6DCP0 A0A1B6E0S4 A0A2P2I6P9 A0A0P6I905
PDB
4DGJ
E-value=6.23819e-28,
Score=306
Ontologies
GO
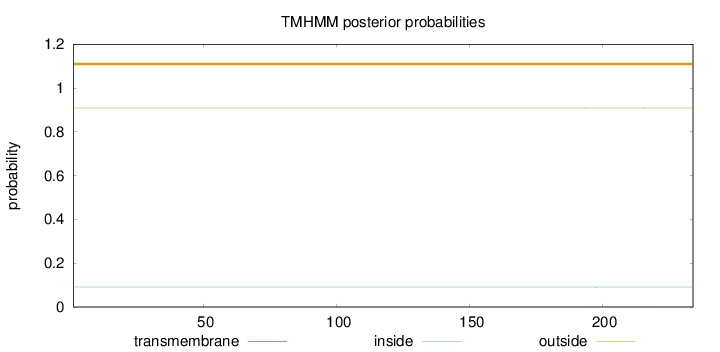
Topology
Subcellular location
Secreted
Length:
234
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01576
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09146
outside
1 - 234
Population Genetic Test Statistics
Pi
59.819166
Theta
48.291836
Tajima's D
-0.782408
CLR
1.131977
CSRT
0.181540922953852
Interpretation
Uncertain
