Gene
KWMTBOMO04171
Pre Gene Modal
BGIBMGA008669
Annotation
PREDICTED:_solute_carrier_organic_anion_transporter_family_member_2B1-like_[Bombyx_mori]
Full name
Solute carrier organic anion transporter family member
Location in the cell
PlasmaMembrane Reliability : 4.95
Sequence
CDS
ATGATGCTACAAGGGAGAACAGTATTTCACAAGTTATGGTACTTCCTCCAAACGTATATCCTGACGCTGAAGAGGTTCGACCTGTTCCTCCAGGGTACCCTGCTTATCGTCATCCTGCTGGAGAGCAACGTGTACTTGCTGCTGAAGAGGGACGCGGACCAGGGTTACAGTACACCTCTTATTGAAAGCTGGGTACAAATCGCAGCAGGTGTCATAGAATGCGTATTGGGAGTTGTTCTGGCGTGGTGGGGGTCTGGTCGACGGCAGTTCGCGCTGACGGGATGGCTAGGAGCCTCCGCTACAGCTGGATTACTGGTCTTAGCATTTCCTTTCGCGTATTCTAATCCTGCGAATGTGGAACTATGCGGGGGTGAACCGGCCTTTCGATCCATAAACCCTAGAGACGAGAACGTGACACCTCGCACCGGGATTCTGGTACTGACGGCGCTGCTATGTGCCCTCACGAAGGTCAGCATCTGGGCGCACGGATTAACCTACTTAGACGACCATGACCCGACCAACGGCCCTTACTTCTATGGTATTCTCATATCGATTCGTTTGTCACTCGGGTTGAGCGGCACCAACTGGTTCCGCCCGAGCTCAGTACGAGACGACTGGTGGGAGGGCCAGCTCTCCGCGTCGATGCTGACGATGATGTTCGCCATTCTCTTCACGTTCTTCCCGACCAAGATGCCACAGTGGCAGGAGGAGGAAGTCACAGTGGATACAGATTTCCTGCCGTCCCTGTATCGAGTGATCAACAATAGAATAGTGGTGATCCAGTCTGTGGCTCTCTCTCTACTCACTACTGCAATCTTCTGCTACGTCGAATACGACCATGCATACGTTCAGGCTCGTTTCCACGTCGAGGCTCTCAGACAGGATCTCAGGACCTCACGATTTTTGAGCGACTTCTTCAGATCGCTGGTCGTCATTTTCTTCATTATGATATTCAGGGTGAGATTCTCAGCGCGTCGTCCTGACGGCGTGAAGGCCAACACGGCGGCTCGAGTGGGCGGCGTGGTGGCTGTCTTCGTGGCCATATTCTTCGTGGTCCTGGCTTCCCTGGGCTGCGACACGGGCACACTCGCGGGGCTGGACGAGGAATACGTTCAGCCAGAGTGCAGTCAGCAATGCGGATGCAACTCGGCTAGATACGGGTTCAGCCCGGTCTGTGGGCTGGACACGCTCACGACGTACTTCTCTCCCTGCCACGCAGGCTGCCGGGCCGTGGAAGACCTTAACGGGTTCTCGTTGTACGCGGACTGCACGTGCGGGGCGCAACGCGCGGCTCTCGGGGCCTGCGCCCTTAGCACGTGCTCCGTGCCCTTCAGTATCTACCAGATATTCTACACTATTATACTTGCAGTCTCAGGAGCATCGTTCCTGATGCAAGGCATGGTGCTAGTGAGGGCGGTCCGCAGGGTCGATAAACCGATCGCGATTGGATTCTCTTTCGCTCTGGTGGCGCTCCTGTCCTTTGTCGTCGGAAGACTACTGTACATGCTCATCAGTTGTAAGTTACCGGTTTCCACATTCGACTCTAGGTTTGAAGAAGACATCGCCCTACCCTGGAACGACCACATAGCCCGAACGTTAACTGCTCATTCTTTAAAAAGAGCGTTTTATGTACTTCCGTAA
Protein
MMLQGRTVFHKLWYFLQTYILTLKRFDLFLQGTLLIVILLESNVYLLLKRDADQGYSTPLIESWVQIAAGVIECVLGVVLAWWGSGRRQFALTGWLGASATAGLLVLAFPFAYSNPANVELCGGEPAFRSINPRDENVTPRTGILVLTALLCALTKVSIWAHGLTYLDDHDPTNGPYFYGILISIRLSLGLSGTNWFRPSSVRDDWWEGQLSASMLTMMFAILFTFFPTKMPQWQEEEVTVDTDFLPSLYRVINNRIVVIQSVALSLLTTAIFCYVEYDHAYVQARFHVEALRQDLRTSRFLSDFFRSLVVIFFIMIFRVRFSARRPDGVKANTAARVGGVVAVFVAIFFVVLASLGCDTGTLAGLDEEYVQPECSQQCGCNSARYGFSPVCGLDTLTTYFSPCHAGCRAVEDLNGFSLYADCTCGAQRAALGACALSTCSVPFSIYQIFYTIILAVSGASFLMQGMVLVRAVRRVDKPIAIGFSFALVALLSFVVGRLLYMLISCKLPVSTFDSRFEEDIALPWNDHIARTLTAHSLKRAFYVLP
Summary
Similarity
Belongs to the organo anion transporter (TC 2.A.60) family.
Uniprot
H9JGM2
A0A2A4JQE9
A0A1E1WNE1
A0A212EPS4
A0A2H1W8B4
A0A194PWF4
+ More
A0A2H1WWI3 A0A212FCG5 A0A194RD13 B4JCP7 A0A194PXB3 A0A0M4E9X9 B4MWM3 A0A3B0J9M8 A0A2J7PEN1 A0A1W4VFG4 Q171D6 W5JS68 A0A182HR41 A0A182JND9 A0A182UKJ1 B3N3F1 A0A182VE03 A0A182MA06 B4G8T8 A0A182GU23 Q7PJX4 B4IE96 A0A0J9TM45 A0A182PDY1 A0A182XHC0 Q9VK82 B4KJ34 B4Q3T8 A0A182JFR2 B3MNT5 A0A182NNV2 A0A182F326 A0A182Y649 A0A0L7R3Z7 A0A182R1S9 A0A1J1HFX6 A0A182W8L3 A0A182Q9P7 A0A1I8M6X7 A0A1A9YND8 A0A1B0AT54 A0A1I8Q241 A0A0L0C5X7 A0A195E3F2 A0A336KHU9 F4WH08 A0A151K0Y9 A0A1A9UH83 A0A0T6AXK3 A0A1W4X3K7 A0A2H8TM63 A0A154P055 A0A336KSL4 A0A2A3E725 A0A232FIW2 A0A034W6D8 A0A2S2PBB3 A0A158NTA4 K7IWN3 A0A088AI24 A0A0K8UY30 E2ABP5 A0A151X5P1 A0A195BUR6 E9INY6 E2BXT4 J9K6L3 W8B1W1
A0A2H1WWI3 A0A212FCG5 A0A194RD13 B4JCP7 A0A194PXB3 A0A0M4E9X9 B4MWM3 A0A3B0J9M8 A0A2J7PEN1 A0A1W4VFG4 Q171D6 W5JS68 A0A182HR41 A0A182JND9 A0A182UKJ1 B3N3F1 A0A182VE03 A0A182MA06 B4G8T8 A0A182GU23 Q7PJX4 B4IE96 A0A0J9TM45 A0A182PDY1 A0A182XHC0 Q9VK82 B4KJ34 B4Q3T8 A0A182JFR2 B3MNT5 A0A182NNV2 A0A182F326 A0A182Y649 A0A0L7R3Z7 A0A182R1S9 A0A1J1HFX6 A0A182W8L3 A0A182Q9P7 A0A1I8M6X7 A0A1A9YND8 A0A1B0AT54 A0A1I8Q241 A0A0L0C5X7 A0A195E3F2 A0A336KHU9 F4WH08 A0A151K0Y9 A0A1A9UH83 A0A0T6AXK3 A0A1W4X3K7 A0A2H8TM63 A0A154P055 A0A336KSL4 A0A2A3E725 A0A232FIW2 A0A034W6D8 A0A2S2PBB3 A0A158NTA4 K7IWN3 A0A088AI24 A0A0K8UY30 E2ABP5 A0A151X5P1 A0A195BUR6 E9INY6 E2BXT4 J9K6L3 W8B1W1
Pubmed
19121390
22118469
26354079
17994087
17510324
20920257
+ More
23761445 26483478 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 25315136 26108605 21719571 28648823 25348373 21347285 20075255 20798317 21282665 24495485
23761445 26483478 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 25315136 26108605 21719571 28648823 25348373 21347285 20075255 20798317 21282665 24495485
EMBL
BABH01034380
BABH01034381
BABH01034382
BABH01034383
NWSH01000785
PCG74221.1
+ More
GDQN01002552 JAT88502.1 AGBW02013393 OWR43493.1 ODYU01006865 SOQ49092.1 KQ459589 KPI97657.1 ODYU01011542 SOQ57347.1 AGBW02009190 OWR51439.1 KQ460367 KPJ15507.1 CH916368 EDW04211.1 KPI97658.1 CP012523 ALC40040.1 CH963857 EDW76512.1 OUUW01000004 SPP78947.1 NEVH01026092 PNF14779.1 CH477459 EAT40590.1 ADMH02000254 ETN67232.1 APCN01001640 CH954177 EDV58791.2 AXCM01000664 CH479180 EDW28768.1 JXUM01002456 JXUM01002457 JXUM01002458 KQ560139 KXJ84224.1 AAAB01008980 EAA43589.4 CH480831 EDW45923.1 CM002910 KMY89925.1 AE014134 AY071008 AAF53197.2 AAL48630.1 CH933807 EDW11396.2 CM000361 EDX04813.1 CH902620 EDV32122.1 KQ414659 KOC65602.1 CVRI01000001 CRK86316.1 AXCN02001989 JXJN01003180 JRES01000869 KNC27652.1 KQ979701 KYN19601.1 UFQS01000438 UFQT01000438 SSX03941.1 SSX24306.1 GL888148 EGI66487.1 KQ981248 KYN44258.1 LJIG01022578 KRT79830.1 GFXV01002947 MBW14752.1 KQ434786 KZC05247.1 SSX03940.1 SSX24305.1 KZ288347 PBC27500.1 NNAY01000156 OXU30470.1 GAKP01009080 JAC49872.1 GGMR01014065 MBY26684.1 ADTU01025770 GDHF01020732 JAI31582.1 GL438334 EFN69147.1 KQ982515 KYQ55609.1 KQ976409 KYM90953.1 GL764397 EFZ17735.1 GL451314 EFN79491.1 ABLF02040023 GAMC01014093 JAB92462.1
GDQN01002552 JAT88502.1 AGBW02013393 OWR43493.1 ODYU01006865 SOQ49092.1 KQ459589 KPI97657.1 ODYU01011542 SOQ57347.1 AGBW02009190 OWR51439.1 KQ460367 KPJ15507.1 CH916368 EDW04211.1 KPI97658.1 CP012523 ALC40040.1 CH963857 EDW76512.1 OUUW01000004 SPP78947.1 NEVH01026092 PNF14779.1 CH477459 EAT40590.1 ADMH02000254 ETN67232.1 APCN01001640 CH954177 EDV58791.2 AXCM01000664 CH479180 EDW28768.1 JXUM01002456 JXUM01002457 JXUM01002458 KQ560139 KXJ84224.1 AAAB01008980 EAA43589.4 CH480831 EDW45923.1 CM002910 KMY89925.1 AE014134 AY071008 AAF53197.2 AAL48630.1 CH933807 EDW11396.2 CM000361 EDX04813.1 CH902620 EDV32122.1 KQ414659 KOC65602.1 CVRI01000001 CRK86316.1 AXCN02001989 JXJN01003180 JRES01000869 KNC27652.1 KQ979701 KYN19601.1 UFQS01000438 UFQT01000438 SSX03941.1 SSX24306.1 GL888148 EGI66487.1 KQ981248 KYN44258.1 LJIG01022578 KRT79830.1 GFXV01002947 MBW14752.1 KQ434786 KZC05247.1 SSX03940.1 SSX24305.1 KZ288347 PBC27500.1 NNAY01000156 OXU30470.1 GAKP01009080 JAC49872.1 GGMR01014065 MBY26684.1 ADTU01025770 GDHF01020732 JAI31582.1 GL438334 EFN69147.1 KQ982515 KYQ55609.1 KQ976409 KYM90953.1 GL764397 EFZ17735.1 GL451314 EFN79491.1 ABLF02040023 GAMC01014093 JAB92462.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000001070
+ More
UP000092553 UP000007798 UP000268350 UP000235965 UP000192221 UP000008820 UP000000673 UP000075840 UP000075881 UP000075902 UP000008711 UP000075903 UP000075883 UP000008744 UP000069940 UP000249989 UP000007062 UP000001292 UP000075885 UP000076407 UP000000803 UP000009192 UP000000304 UP000075880 UP000007801 UP000075884 UP000069272 UP000076408 UP000053825 UP000075900 UP000183832 UP000075920 UP000075886 UP000095301 UP000092443 UP000092460 UP000095300 UP000037069 UP000078492 UP000007755 UP000078541 UP000078200 UP000192223 UP000076502 UP000242457 UP000215335 UP000005205 UP000002358 UP000005203 UP000000311 UP000075809 UP000078540 UP000008237 UP000007819
UP000092553 UP000007798 UP000268350 UP000235965 UP000192221 UP000008820 UP000000673 UP000075840 UP000075881 UP000075902 UP000008711 UP000075903 UP000075883 UP000008744 UP000069940 UP000249989 UP000007062 UP000001292 UP000075885 UP000076407 UP000000803 UP000009192 UP000000304 UP000075880 UP000007801 UP000075884 UP000069272 UP000076408 UP000053825 UP000075900 UP000183832 UP000075920 UP000075886 UP000095301 UP000092443 UP000092460 UP000095300 UP000037069 UP000078492 UP000007755 UP000078541 UP000078200 UP000192223 UP000076502 UP000242457 UP000215335 UP000005205 UP000002358 UP000005203 UP000000311 UP000075809 UP000078540 UP000008237 UP000007819
Interpro
CDD
ProteinModelPortal
H9JGM2
A0A2A4JQE9
A0A1E1WNE1
A0A212EPS4
A0A2H1W8B4
A0A194PWF4
+ More
A0A2H1WWI3 A0A212FCG5 A0A194RD13 B4JCP7 A0A194PXB3 A0A0M4E9X9 B4MWM3 A0A3B0J9M8 A0A2J7PEN1 A0A1W4VFG4 Q171D6 W5JS68 A0A182HR41 A0A182JND9 A0A182UKJ1 B3N3F1 A0A182VE03 A0A182MA06 B4G8T8 A0A182GU23 Q7PJX4 B4IE96 A0A0J9TM45 A0A182PDY1 A0A182XHC0 Q9VK82 B4KJ34 B4Q3T8 A0A182JFR2 B3MNT5 A0A182NNV2 A0A182F326 A0A182Y649 A0A0L7R3Z7 A0A182R1S9 A0A1J1HFX6 A0A182W8L3 A0A182Q9P7 A0A1I8M6X7 A0A1A9YND8 A0A1B0AT54 A0A1I8Q241 A0A0L0C5X7 A0A195E3F2 A0A336KHU9 F4WH08 A0A151K0Y9 A0A1A9UH83 A0A0T6AXK3 A0A1W4X3K7 A0A2H8TM63 A0A154P055 A0A336KSL4 A0A2A3E725 A0A232FIW2 A0A034W6D8 A0A2S2PBB3 A0A158NTA4 K7IWN3 A0A088AI24 A0A0K8UY30 E2ABP5 A0A151X5P1 A0A195BUR6 E9INY6 E2BXT4 J9K6L3 W8B1W1
A0A2H1WWI3 A0A212FCG5 A0A194RD13 B4JCP7 A0A194PXB3 A0A0M4E9X9 B4MWM3 A0A3B0J9M8 A0A2J7PEN1 A0A1W4VFG4 Q171D6 W5JS68 A0A182HR41 A0A182JND9 A0A182UKJ1 B3N3F1 A0A182VE03 A0A182MA06 B4G8T8 A0A182GU23 Q7PJX4 B4IE96 A0A0J9TM45 A0A182PDY1 A0A182XHC0 Q9VK82 B4KJ34 B4Q3T8 A0A182JFR2 B3MNT5 A0A182NNV2 A0A182F326 A0A182Y649 A0A0L7R3Z7 A0A182R1S9 A0A1J1HFX6 A0A182W8L3 A0A182Q9P7 A0A1I8M6X7 A0A1A9YND8 A0A1B0AT54 A0A1I8Q241 A0A0L0C5X7 A0A195E3F2 A0A336KHU9 F4WH08 A0A151K0Y9 A0A1A9UH83 A0A0T6AXK3 A0A1W4X3K7 A0A2H8TM63 A0A154P055 A0A336KSL4 A0A2A3E725 A0A232FIW2 A0A034W6D8 A0A2S2PBB3 A0A158NTA4 K7IWN3 A0A088AI24 A0A0K8UY30 E2ABP5 A0A151X5P1 A0A195BUR6 E9INY6 E2BXT4 J9K6L3 W8B1W1
Ontologies
KEGG
GO
PANTHER
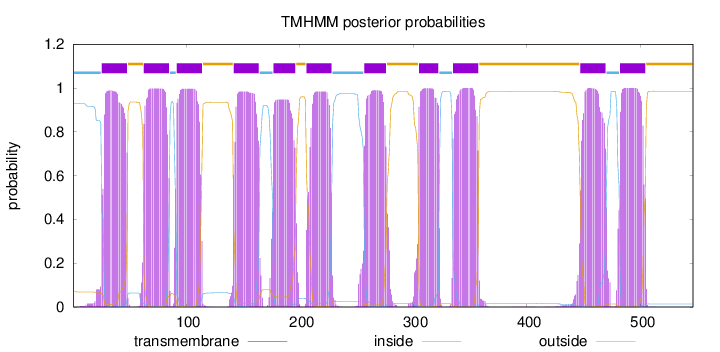
Topology
Subcellular location
Cell membrane
Length:
546
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
235.52062
Exp number, first 60 AAs:
21.50879
Total prob of N-in:
0.93081
POSSIBLE N-term signal
sequence
inside
1 - 25
TMhelix
26 - 48
outside
49 - 62
TMhelix
63 - 85
inside
86 - 91
TMhelix
92 - 114
outside
115 - 141
TMhelix
142 - 164
inside
165 - 176
TMhelix
177 - 196
outside
197 - 205
TMhelix
206 - 228
inside
229 - 256
TMhelix
257 - 276
outside
277 - 304
TMhelix
305 - 322
inside
323 - 334
TMhelix
335 - 357
outside
358 - 446
TMhelix
447 - 469
inside
470 - 481
TMhelix
482 - 504
outside
505 - 546
Population Genetic Test Statistics
Pi
231.230863
Theta
163.664011
Tajima's D
1.274161
CLR
0.073403
CSRT
0.729913504324784
Interpretation
Uncertain
