Gene
KWMTBOMO04170 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008725
Annotation
PREDICTED:_UTP--glucose-1-phosphate_uridylyltransferase_isoform_X2_[Bombyx_mori]
Full name
UTP--glucose-1-phosphate uridylyltransferase
Location in the cell
Cytoplasmic Reliability : 2.174 Nuclear Reliability : 1.732
Sequence
CDS
ATGAAGCCAGAAACCAACGACTCCGATAAAAAGATTCGCAGTCACCAGCGGACGCCGTCAGGGTCTCGTGACTTCAAGGAGGCCACCAAACGTGATGCCCTCGCTCGACTGGAGGTGGAGCTGGAGAAGCTCATCTCCACGGCCCCGGAAACCAGGCGTCCGCAGCTCGAGAAAGAGTTCAAAGGCTTCAAGACGCTCTTCAGCAGATTCTTGGCTGAACAGGGTCCGTCGGTAACATGGGAGAAAATCGAGAAGCTTCCAGAAGGAGCTGTAATCGATTATAACAGTCTGTCCACGCCAACGACGGACAACGTGCACCATATGCTCGACAAGTTGGTGGTGGTGAAGCTTAACGGAGGCCTGGGTACATCCATGGGCTGTAAGGGCCCCAAATCGGTGATCCAAGTCCGGAATGACCTGACCTTCCTTGATCTTACCGTACAGCAAATAGAGCACTTGAACAAGACGTATAAATGCAACGTCCCACTAGTCCTGATGAACTCCTTCAACACGGATGAAGACACCCAGAAGGTGATCAGGAAGTATAAGGGTCTGAAGCTGGACATCCACACCTTCAATCAGTCCTGCCACCCTCGTATTAACAGGGAGTCACTATTGCCCATCGCTCAGAACGGTGATGTTCAAGACGATATTGAATCCTGGTATCCACCGGGTCACGGAGACTTCTACGAGTCATTCCACAATTCCGGTCTCCTACAGAAGTTCATCAACGAGGGTCGTACCTATTGCTTCATCAGCAACATCGACAACTTAGGAGCCACCGTTGACCTCAACATCTTAAGCCTGCTCCTCAACCCGGATCCTCTGCGGCCGGTTCCGGAGTTCGTCATGGAAGTGACCGACAAAACCCGTGCCGACGTCAAGGGAGGAACCCTCATACAATACGAAGACAAATTGCGGCTTTTAGAAATCGCCCAAGTGCCCAAAGAACGCGTTGACGATTTTAAATCTGTTAGCCAATTCAAGTTTTTCAACACGAACAACCTTTGGGCCAAATTGGACGCAGTTCAGCGGGTGGTCGACCAGGGGAGCCTCAACATGGAAATCATTGTCAACAACAAGCATCTATCGGACGGTATCAATGTGATTCAACTAGAGACGGCTGTGGGGGCCGCCATGAAGTGCTTCGAGGGTGGGATCGGCGTCAACGTGCCCCGCAGCAGGTTCTTGCCGGTCAAAAAGACCTCGGACTTGCTGTTGGTCATGTCCAACCTGTACAGCTTGTCCCACGGGTCCTTAGTCATGTCCTCTCAGAGGATGTTTGCCTCGACCCCCCTCGTCAAACTAGGAGACAATCATTTCGCTAAAGTCAAAGAGTACCTGAACAGGTTCGCGACCATACCGGATCTGATCGAACTGGACCATTTGACCGTTTCTGGCGACGTGACCTTCGGAAGAGGAGTGTCTTTGAAGGGCACAGTGATCATAATCGCGAATCACGGCGACCGTATCGACATCCCATCGGGTGCGATTCTCGAAAACAAAATCGTTTCTGGAAACTTAAGGATTTTGGATCACTGA
Protein
MKPETNDSDKKIRSHQRTPSGSRDFKEATKRDALARLEVELEKLISTAPETRRPQLEKEFKGFKTLFSRFLAEQGPSVTWEKIEKLPEGAVIDYNSLSTPTTDNVHHMLDKLVVVKLNGGLGTSMGCKGPKSVIQVRNDLTFLDLTVQQIEHLNKTYKCNVPLVLMNSFNTDEDTQKVIRKYKGLKLDIHTFNQSCHPRINRESLLPIAQNGDVQDDIESWYPPGHGDFYESFHNSGLLQKFINEGRTYCFISNIDNLGATVDLNILSLLLNPDPLRPVPEFVMEVTDKTRADVKGGTLIQYEDKLRLLEIAQVPKERVDDFKSVSQFKFFNTNNLWAKLDAVQRVVDQGSLNMEIIVNNKHLSDGINVIQLETAVGAAMKCFEGGIGVNVPRSRFLPVKKTSDLLLVMSNLYSLSHGSLVMSSQRMFASTPLVKLGDNHFAKVKEYLNRFATIPDLIELDHLTVSGDVTFGRGVSLKGTVIIIANHGDRIDIPSGAILENKIVSGNLRILDH
Summary
Catalytic Activity
alpha-D-glucose 1-phosphate + H(+) + UTP = diphosphate + UDP-alpha-D-glucose
Similarity
Belongs to the UDPGP type 1 family.
Uniprot
H9JGS8
A0A194RHU0
A0A194PWA7
E1U7I5
A0A2W1BUV6
A0A0S1MMM3
+ More
A0A1E1W5G4 A0A2A4JRX6 A0A2A4JQ81 A0A2A4JRI8 A0A2A4JS45 A0A1E1WVB3 A0A2H1W7T1 A0A1E1W9C6 A0A1E1WHG6 A0A3S2TQR1 A0A212EPR9 A0A0T6AST8 D6WPU3 A0A2J7R1R8 A0A2J7R1S7 A0A2J7R1R4 A0A1Y1LAB2 A0A067QYU1 A0A1B6CPT6 A0A1B6CUM9 A0A1B6DXY4 A0A1Y1LF18 T1DE51 A0A023EUR8 A0A1B6F8J5 A0A1B6GT97 A0A182JBG7 A0A1Q3FLA2 A0A182GHQ7 A0A1B6EKX9 A0A182RA51 A0A182QQM1 A0A1B6FUC7 A0A1Y9H383 B0WFJ8 A0A2M4BKP7 A0A1S4FB16 A0A182YQ17 Q58I82 A0A026W0X2 A0A1Y9H2U2 A0A182JS08 A0A1Q3FLG7 T1P8F9 A0A182PRS3 A0A1L8E255 A0A1Y9IVL5 W5J6Z2 T1PNE3 A0A1Y9IZZ0 A0A182LZ51 A0A182HKF0 A0A1W4WHM5 A0A2M4A3B4 A0A2M4CX13 A0A3L8DN04 A0A182TZX8 A0A2M3ZFJ9 F5HMV1 A0A1W4W669 A0A2M4A391 A0A1Y9J0B5 A0A1I8N4L5 A0A1B0CVE3 A0A034VM77 Q7PKK3 A0A1W4WGL4 D3TMX2 A0A0K8V8C7 W8B5P2 A0A1I8N4K5 A0A034VN84 A0A182UQK7 A0A0K8V683 A0A0K8WFB0 A0A023EW50 A0A1B6M968 A0A195F1K1 A0A224XEQ3 E9J656 A0A2A3E8J6 V9ICG4 A0A0Q9WSJ1 B4IXV6 A0A0Q9XET6 A0A195B242 A0A1B6MUL7 A0A0Q9XPW8 A0A1B6LL61 A0A1I8NW79 A0A158NGD1 A0A0Q9X342 E1ZYZ4 A0A1B0A749
A0A1E1W5G4 A0A2A4JRX6 A0A2A4JQ81 A0A2A4JRI8 A0A2A4JS45 A0A1E1WVB3 A0A2H1W7T1 A0A1E1W9C6 A0A1E1WHG6 A0A3S2TQR1 A0A212EPR9 A0A0T6AST8 D6WPU3 A0A2J7R1R8 A0A2J7R1S7 A0A2J7R1R4 A0A1Y1LAB2 A0A067QYU1 A0A1B6CPT6 A0A1B6CUM9 A0A1B6DXY4 A0A1Y1LF18 T1DE51 A0A023EUR8 A0A1B6F8J5 A0A1B6GT97 A0A182JBG7 A0A1Q3FLA2 A0A182GHQ7 A0A1B6EKX9 A0A182RA51 A0A182QQM1 A0A1B6FUC7 A0A1Y9H383 B0WFJ8 A0A2M4BKP7 A0A1S4FB16 A0A182YQ17 Q58I82 A0A026W0X2 A0A1Y9H2U2 A0A182JS08 A0A1Q3FLG7 T1P8F9 A0A182PRS3 A0A1L8E255 A0A1Y9IVL5 W5J6Z2 T1PNE3 A0A1Y9IZZ0 A0A182LZ51 A0A182HKF0 A0A1W4WHM5 A0A2M4A3B4 A0A2M4CX13 A0A3L8DN04 A0A182TZX8 A0A2M3ZFJ9 F5HMV1 A0A1W4W669 A0A2M4A391 A0A1Y9J0B5 A0A1I8N4L5 A0A1B0CVE3 A0A034VM77 Q7PKK3 A0A1W4WGL4 D3TMX2 A0A0K8V8C7 W8B5P2 A0A1I8N4K5 A0A034VN84 A0A182UQK7 A0A0K8V683 A0A0K8WFB0 A0A023EW50 A0A1B6M968 A0A195F1K1 A0A224XEQ3 E9J656 A0A2A3E8J6 V9ICG4 A0A0Q9WSJ1 B4IXV6 A0A0Q9XET6 A0A195B242 A0A1B6MUL7 A0A0Q9XPW8 A0A1B6LL61 A0A1I8NW79 A0A158NGD1 A0A0Q9X342 E1ZYZ4 A0A1B0A749
EC Number
2.7.7.9
Pubmed
EMBL
BABH01034380
KQ460367
KPJ15511.1
KQ459589
KPI97661.1
FJ793042
+ More
ACY69181.1 KZ149929 PZC77445.1 KR821075 ALL42062.1 GDQN01008827 JAT82227.1 NWSH01000785 PCG74223.1 PCG74225.1 PCG74224.1 PCG74222.1 GDQN01000136 JAT90918.1 ODYU01006865 SOQ49093.1 GDQN01007480 JAT83574.1 GDQN01004646 JAT86408.1 RSAL01000020 RVE52571.1 AGBW02013393 OWR43492.1 LJIG01022885 KRT78224.1 KQ971354 EFA06874.2 NEVH01008202 PNF34777.1 PNF34778.1 PNF34779.1 GEZM01061319 JAV70564.1 KK852818 KDR15697.1 GEDC01021796 JAS15502.1 GEDC01020365 JAS16933.1 GEDC01006762 JAS30536.1 GEZM01061317 JAV70565.1 GALA01001187 JAA93665.1 GAPW01000876 JAC12722.1 GECZ01023222 JAS46547.1 GECZ01004117 JAS65652.1 GFDL01006698 JAV28347.1 JXUM01064342 KQ562293 KXJ76222.1 GECZ01031181 JAS38588.1 AXCN02001509 GECZ01015942 JAS53827.1 DS231918 EDS26266.1 GGFJ01004390 MBW53531.1 AY944757 CH477349 AAX47080.1 EAT42894.1 KK107533 EZA49231.1 GFDL01006658 JAV28387.1 KA644934 AFP59563.1 GFDF01001365 JAV12719.1 ADMH02002101 ETN59168.1 KA649166 KA649590 AFP64219.1 AXCM01000640 GGFK01001928 MBW35249.1 GGFL01005688 MBW69866.1 QOIP01000006 RLU21810.1 GGFM01006573 MBW27324.1 AAAB01008987 EGK97623.1 GGFK01001869 MBW35190.1 AJWK01030438 GAKP01015373 GAKP01015367 JAC43579.1 EAA43227.3 EZ422774 ADD19050.1 GDHF01017157 JAI35157.1 GAMC01021461 JAB85094.1 GAKP01015371 GAKP01015369 GAKP01015365 GAKP01015363 JAC43587.1 GDHF01018199 GDHF01003124 JAI34115.1 JAI49190.1 GDHF01002463 JAI49851.1 GAPW01001009 JAC12589.1 GEBQ01007507 JAT32470.1 KQ981864 KYN34256.1 GFTR01006942 JAW09484.1 GL768221 EFZ11766.1 KZ288349 PBC27381.1 JR039603 AEY58775.1 CH964168 KRF99230.1 CH916366 EDV96477.1 CH933809 KRG06169.1 KQ976662 KYM78546.1 GEBQ01023084 GEBQ01000395 JAT16893.1 JAT39582.1 KRG06168.1 GEBQ01015505 JAT24472.1 ADTU01014827 ADTU01014828 KRF99232.1 GL435242 EFN73684.1
ACY69181.1 KZ149929 PZC77445.1 KR821075 ALL42062.1 GDQN01008827 JAT82227.1 NWSH01000785 PCG74223.1 PCG74225.1 PCG74224.1 PCG74222.1 GDQN01000136 JAT90918.1 ODYU01006865 SOQ49093.1 GDQN01007480 JAT83574.1 GDQN01004646 JAT86408.1 RSAL01000020 RVE52571.1 AGBW02013393 OWR43492.1 LJIG01022885 KRT78224.1 KQ971354 EFA06874.2 NEVH01008202 PNF34777.1 PNF34778.1 PNF34779.1 GEZM01061319 JAV70564.1 KK852818 KDR15697.1 GEDC01021796 JAS15502.1 GEDC01020365 JAS16933.1 GEDC01006762 JAS30536.1 GEZM01061317 JAV70565.1 GALA01001187 JAA93665.1 GAPW01000876 JAC12722.1 GECZ01023222 JAS46547.1 GECZ01004117 JAS65652.1 GFDL01006698 JAV28347.1 JXUM01064342 KQ562293 KXJ76222.1 GECZ01031181 JAS38588.1 AXCN02001509 GECZ01015942 JAS53827.1 DS231918 EDS26266.1 GGFJ01004390 MBW53531.1 AY944757 CH477349 AAX47080.1 EAT42894.1 KK107533 EZA49231.1 GFDL01006658 JAV28387.1 KA644934 AFP59563.1 GFDF01001365 JAV12719.1 ADMH02002101 ETN59168.1 KA649166 KA649590 AFP64219.1 AXCM01000640 GGFK01001928 MBW35249.1 GGFL01005688 MBW69866.1 QOIP01000006 RLU21810.1 GGFM01006573 MBW27324.1 AAAB01008987 EGK97623.1 GGFK01001869 MBW35190.1 AJWK01030438 GAKP01015373 GAKP01015367 JAC43579.1 EAA43227.3 EZ422774 ADD19050.1 GDHF01017157 JAI35157.1 GAMC01021461 JAB85094.1 GAKP01015371 GAKP01015369 GAKP01015365 GAKP01015363 JAC43587.1 GDHF01018199 GDHF01003124 JAI34115.1 JAI49190.1 GDHF01002463 JAI49851.1 GAPW01001009 JAC12589.1 GEBQ01007507 JAT32470.1 KQ981864 KYN34256.1 GFTR01006942 JAW09484.1 GL768221 EFZ11766.1 KZ288349 PBC27381.1 JR039603 AEY58775.1 CH964168 KRF99230.1 CH916366 EDV96477.1 CH933809 KRG06169.1 KQ976662 KYM78546.1 GEBQ01023084 GEBQ01000395 JAT16893.1 JAT39582.1 KRG06168.1 GEBQ01015505 JAT24472.1 ADTU01014827 ADTU01014828 KRF99232.1 GL435242 EFN73684.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000007266 UP000235965 UP000027135 UP000075880 UP000069940 UP000249989 UP000075900 UP000075886 UP000075884 UP000002320 UP000076408 UP000008820 UP000053097 UP000075881 UP000075885 UP000075920 UP000000673 UP000095301 UP000076407 UP000075883 UP000192223 UP000279307 UP000075902 UP000007062 UP000092461 UP000075903 UP000078541 UP000242457 UP000007798 UP000001070 UP000009192 UP000078540 UP000095300 UP000005205 UP000000311 UP000092445
UP000007266 UP000235965 UP000027135 UP000075880 UP000069940 UP000249989 UP000075900 UP000075886 UP000075884 UP000002320 UP000076408 UP000008820 UP000053097 UP000075881 UP000075885 UP000075920 UP000000673 UP000095301 UP000076407 UP000075883 UP000192223 UP000279307 UP000075902 UP000007062 UP000092461 UP000075903 UP000078541 UP000242457 UP000007798 UP000001070 UP000009192 UP000078540 UP000095300 UP000005205 UP000000311 UP000092445
PRIDE
Interpro
IPR016267
UDPGP_trans
+ More
IPR002618 UDPGP_fam
IPR029044 Nucleotide-diphossugar_trans
IPR002891 APS_kinase
IPR027417 P-loop_NTPase
IPR025980 ATP-Sase_PUA-like_dom
IPR002650 Sulphate_adenylyltransferase
IPR012934 Znf_AD
IPR031632 SVIP
IPR024951 Sulfurylase_cat_dom
IPR015947 PUA-like_sf
IPR014729 Rossmann-like_a/b/a_fold
IPR002618 UDPGP_fam
IPR029044 Nucleotide-diphossugar_trans
IPR002891 APS_kinase
IPR027417 P-loop_NTPase
IPR025980 ATP-Sase_PUA-like_dom
IPR002650 Sulphate_adenylyltransferase
IPR012934 Znf_AD
IPR031632 SVIP
IPR024951 Sulfurylase_cat_dom
IPR015947 PUA-like_sf
IPR014729 Rossmann-like_a/b/a_fold
Gene 3D
ProteinModelPortal
H9JGS8
A0A194RHU0
A0A194PWA7
E1U7I5
A0A2W1BUV6
A0A0S1MMM3
+ More
A0A1E1W5G4 A0A2A4JRX6 A0A2A4JQ81 A0A2A4JRI8 A0A2A4JS45 A0A1E1WVB3 A0A2H1W7T1 A0A1E1W9C6 A0A1E1WHG6 A0A3S2TQR1 A0A212EPR9 A0A0T6AST8 D6WPU3 A0A2J7R1R8 A0A2J7R1S7 A0A2J7R1R4 A0A1Y1LAB2 A0A067QYU1 A0A1B6CPT6 A0A1B6CUM9 A0A1B6DXY4 A0A1Y1LF18 T1DE51 A0A023EUR8 A0A1B6F8J5 A0A1B6GT97 A0A182JBG7 A0A1Q3FLA2 A0A182GHQ7 A0A1B6EKX9 A0A182RA51 A0A182QQM1 A0A1B6FUC7 A0A1Y9H383 B0WFJ8 A0A2M4BKP7 A0A1S4FB16 A0A182YQ17 Q58I82 A0A026W0X2 A0A1Y9H2U2 A0A182JS08 A0A1Q3FLG7 T1P8F9 A0A182PRS3 A0A1L8E255 A0A1Y9IVL5 W5J6Z2 T1PNE3 A0A1Y9IZZ0 A0A182LZ51 A0A182HKF0 A0A1W4WHM5 A0A2M4A3B4 A0A2M4CX13 A0A3L8DN04 A0A182TZX8 A0A2M3ZFJ9 F5HMV1 A0A1W4W669 A0A2M4A391 A0A1Y9J0B5 A0A1I8N4L5 A0A1B0CVE3 A0A034VM77 Q7PKK3 A0A1W4WGL4 D3TMX2 A0A0K8V8C7 W8B5P2 A0A1I8N4K5 A0A034VN84 A0A182UQK7 A0A0K8V683 A0A0K8WFB0 A0A023EW50 A0A1B6M968 A0A195F1K1 A0A224XEQ3 E9J656 A0A2A3E8J6 V9ICG4 A0A0Q9WSJ1 B4IXV6 A0A0Q9XET6 A0A195B242 A0A1B6MUL7 A0A0Q9XPW8 A0A1B6LL61 A0A1I8NW79 A0A158NGD1 A0A0Q9X342 E1ZYZ4 A0A1B0A749
A0A1E1W5G4 A0A2A4JRX6 A0A2A4JQ81 A0A2A4JRI8 A0A2A4JS45 A0A1E1WVB3 A0A2H1W7T1 A0A1E1W9C6 A0A1E1WHG6 A0A3S2TQR1 A0A212EPR9 A0A0T6AST8 D6WPU3 A0A2J7R1R8 A0A2J7R1S7 A0A2J7R1R4 A0A1Y1LAB2 A0A067QYU1 A0A1B6CPT6 A0A1B6CUM9 A0A1B6DXY4 A0A1Y1LF18 T1DE51 A0A023EUR8 A0A1B6F8J5 A0A1B6GT97 A0A182JBG7 A0A1Q3FLA2 A0A182GHQ7 A0A1B6EKX9 A0A182RA51 A0A182QQM1 A0A1B6FUC7 A0A1Y9H383 B0WFJ8 A0A2M4BKP7 A0A1S4FB16 A0A182YQ17 Q58I82 A0A026W0X2 A0A1Y9H2U2 A0A182JS08 A0A1Q3FLG7 T1P8F9 A0A182PRS3 A0A1L8E255 A0A1Y9IVL5 W5J6Z2 T1PNE3 A0A1Y9IZZ0 A0A182LZ51 A0A182HKF0 A0A1W4WHM5 A0A2M4A3B4 A0A2M4CX13 A0A3L8DN04 A0A182TZX8 A0A2M3ZFJ9 F5HMV1 A0A1W4W669 A0A2M4A391 A0A1Y9J0B5 A0A1I8N4L5 A0A1B0CVE3 A0A034VM77 Q7PKK3 A0A1W4WGL4 D3TMX2 A0A0K8V8C7 W8B5P2 A0A1I8N4K5 A0A034VN84 A0A182UQK7 A0A0K8V683 A0A0K8WFB0 A0A023EW50 A0A1B6M968 A0A195F1K1 A0A224XEQ3 E9J656 A0A2A3E8J6 V9ICG4 A0A0Q9WSJ1 B4IXV6 A0A0Q9XET6 A0A195B242 A0A1B6MUL7 A0A0Q9XPW8 A0A1B6LL61 A0A1I8NW79 A0A158NGD1 A0A0Q9X342 E1ZYZ4 A0A1B0A749
PDB
4R7P
E-value=0,
Score=1658
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
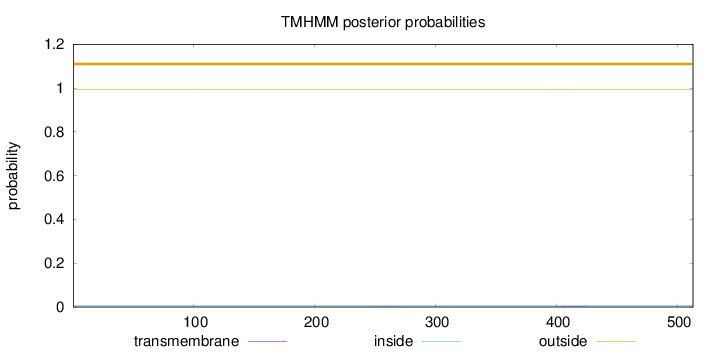
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0358200000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00599
outside
1 - 513
Population Genetic Test Statistics
Pi
267.831313
Theta
171.254329
Tajima's D
1.830963
CLR
0.044444
CSRT
0.851757412129393
Interpretation
Uncertain
