Gene
KWMTBOMO04165
Pre Gene Modal
BGIBMGA008671
Annotation
PREDICTED:_cyclin-H_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.2
Sequence
CDS
ATGTTTTCTACCAGCTCTCAAAGAAAATTTTGGACGTTCAGTGATGAAAATGAACTCGCACGCTTACGTGAAAAACACAATTATTTTTTTTACGCGAGACAGAACTCGCACATCGACGAGCAACAACGTTTCACGTATTTCTTGTCTCCTGATGAAGAACGACAACTTTTAAAGCAATACGAACTCCATCTGAAAGAATTCTGCAAGAGATTCACACCTCCAATGCCCAAAGGGGTGGTGGGCACAGCCTTTCATTACTTTAAACGATTTTATCTATACAACTCGACAATGGACTATCATCCAAAAGAAATTTTAGCCACTTGCGTGTATTTAGCGTGTAAAGTTGAAGAATTTAACGTTTCTATCGGGCAATTCGTCGCCAATATCAAAGGAGATCGAGAAAAAGCATCAGATATAATACTAAACAATGAATTATTGCTAATGCAACAATTAAACTACCATTTGACCATACATAATCCTTTCAGACCAGTAGAAGGATTCCTAATAGACATTAAAACTAGATGTTCATTGGCTAATCCTGAACGTCTTCGTAGTGGTATTGATGAGTTCCTAGAAAAGGTGTTCTTAACTGATGCATGCTTGTTGTATGCTCCGTCACAAATTGCTTTAGCTGCAGTACTCCATGCAGCGAGCAAGGAACAAGAGAACTTGGATAGCTACGTGACAGACATACTCTTCAGAGATGCGGGACGTGATAAATTGGCGATACTTATTGAAGCTGTAAGAAAGATTCGATCATTAGTAAAAATGGTAGAGGCACCAGCTAGAGAACGAGTACGGCTTATAGAGAAGAAATTAGACAAATGTCGGAACCAAGAAAATAACCCTGACAGTGAGATTTACAAGCGTCGGATGAGAGAGCTGCTAGACGAAGACGATGTTTTACATGTAGGATCCAGTAGAATGTCTTTAGATACAAGTGGAGTTACACAAGTTCTATCACCTTCTGCCTCTTAA
Protein
MFSTSSQRKFWTFSDENELARLREKHNYFFYARQNSHIDEQQRFTYFLSPDEERQLLKQYELHLKEFCKRFTPPMPKGVVGTAFHYFKRFYLYNSTMDYHPKEILATCVYLACKVEEFNVSIGQFVANIKGDREKASDIILNNELLLMQQLNYHLTIHNPFRPVEGFLIDIKTRCSLANPERLRSGIDEFLEKVFLTDACLLYAPSQIALAAVLHAASKEQENLDSYVTDILFRDAGRDKLAILIEAVRKIRSLVKMVEAPARERVRLIEKKLDKCRNQENNPDSEIYKRRMRELLDEDDVLHVGSSRMSLDTSGVTQVLSPSAS
Summary
Similarity
Belongs to the cyclin family.
Belongs to the AKH/HRTH/RPCH family.
Belongs to the AKH/HRTH/RPCH family.
Uniprot
H9JGM4
A0A2H1WI10
A0A2W1BQZ3
A0A2A4JQZ1
A0A194PWA2
A0A0L7KWS8
+ More
S4PMU2 A0A3S2PIR6 A0A212FKR7 A0A1E1VYT2 A0A194RDG2 Q16JQ2 A0A182UXN1 A0A182L0E9 Q7Q4I7 A0A182IC32 A0A182GYI0 A0A182UFH5 A0A2M4BTG6 A0A2M4BTE8 A0A182XFU7 A0A182K6S6 W5J9W4 A0A182SFC9 A0A182WHC3 A0A182Y9Y1 A0A182RG34 A0A182NET9 A0A182F5K1 A0A182PAP1 A0A182QFH8 A0A182IJV9 W8AQE1 A0A034WGT5 B0WCS0 A0A1Q3F1P6 A0A182M243 A0A336MBU1 U5EZI0 A0A336MAL8 A0A084VMA4 A0A0A1WNN0 A0A0K8TL95 A0A0K8UW44 A0A2P8XMK8 A0A1I8MZY5 A0A0J7LA15 A0A3L8DIR2 D3TRT0 A0A1B0G766 A0A1A9ZXB0 F4X0Q1 A0A158NCY3 E9IJX2 A0A195F817 A0A154P750 A0A151WYF4 A0A195D1R5 A0A0N0BEY2 A0A2J7Q483 A0A151J3D1 E2ASF4 A0A087ZN02 A0A067QJI2 A0A1A9UQ25 A0A0L7R4B0 A0A1Y1LP72 A0A0L0CF97 E2BY23 A0A1A9YB41 A0A2A3EIF7 A0A1W4WY47 A0A1B0DB14 K7IUX2 Q2M0B7 B3MAT4 B4LCT9 A0A3B0JTX4 A0A232EZX0 A0A1W4USX4 B4KUV3 A0A0C9QHN8 B4MM61 N6TWG3 U4UCW3 B4IAW2 B4GRV0 B4ITC1 B4IZ94 B4QKN5 D6WRD7 B3NJ00 O76513 C6SV35 Q32KG2 O17144 A0A1A9WQR9 A0A1L8DWS4 A0A1B6LS70 A0A1S4ESS3
S4PMU2 A0A3S2PIR6 A0A212FKR7 A0A1E1VYT2 A0A194RDG2 Q16JQ2 A0A182UXN1 A0A182L0E9 Q7Q4I7 A0A182IC32 A0A182GYI0 A0A182UFH5 A0A2M4BTG6 A0A2M4BTE8 A0A182XFU7 A0A182K6S6 W5J9W4 A0A182SFC9 A0A182WHC3 A0A182Y9Y1 A0A182RG34 A0A182NET9 A0A182F5K1 A0A182PAP1 A0A182QFH8 A0A182IJV9 W8AQE1 A0A034WGT5 B0WCS0 A0A1Q3F1P6 A0A182M243 A0A336MBU1 U5EZI0 A0A336MAL8 A0A084VMA4 A0A0A1WNN0 A0A0K8TL95 A0A0K8UW44 A0A2P8XMK8 A0A1I8MZY5 A0A0J7LA15 A0A3L8DIR2 D3TRT0 A0A1B0G766 A0A1A9ZXB0 F4X0Q1 A0A158NCY3 E9IJX2 A0A195F817 A0A154P750 A0A151WYF4 A0A195D1R5 A0A0N0BEY2 A0A2J7Q483 A0A151J3D1 E2ASF4 A0A087ZN02 A0A067QJI2 A0A1A9UQ25 A0A0L7R4B0 A0A1Y1LP72 A0A0L0CF97 E2BY23 A0A1A9YB41 A0A2A3EIF7 A0A1W4WY47 A0A1B0DB14 K7IUX2 Q2M0B7 B3MAT4 B4LCT9 A0A3B0JTX4 A0A232EZX0 A0A1W4USX4 B4KUV3 A0A0C9QHN8 B4MM61 N6TWG3 U4UCW3 B4IAW2 B4GRV0 B4ITC1 B4IZ94 B4QKN5 D6WRD7 B3NJ00 O76513 C6SV35 Q32KG2 O17144 A0A1A9WQR9 A0A1L8DWS4 A0A1B6LS70 A0A1S4ESS3
Pubmed
19121390
28756777
26354079
26227816
23622113
22118469
+ More
17510324 20966253 12364791 14747013 17210077 26483478 20920257 23761445 25244985 24495485 25348373 24438588 25830018 26369729 29403074 25315136 30249741 20353571 21719571 21347285 21282665 20798317 24845553 28004739 26108605 20075255 15632085 17994087 28648823 23537049 17550304 22936249 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
17510324 20966253 12364791 14747013 17210077 26483478 20920257 23761445 25244985 24495485 25348373 24438588 25830018 26369729 29403074 25315136 30249741 20353571 21719571 21347285 21282665 20798317 24845553 28004739 26108605 20075255 15632085 17994087 28648823 23537049 17550304 22936249 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01034378
ODYU01008799
SOQ52710.1
KZ149929
PZC77448.1
NWSH01000785
+ More
PCG74229.1 KQ459589 KPI97656.1 JTDY01004984 KOB67509.1 GAIX01003380 JAA89180.1 RSAL01000020 RVE52576.1 AGBW02007996 OWR54334.1 GDQN01011169 JAT79885.1 KQ460367 KPJ15504.1 CH477997 EAT34507.1 EAT34510.1 AAAB01008964 EAA12839.3 APCN01000774 JXUM01097524 KQ564351 KXJ72389.1 GGFJ01007226 MBW56367.1 GGFJ01007225 MBW56366.1 ADMH02001866 ETN60786.1 AXCN02000840 GAMC01015547 GAMC01015546 GAMC01015545 JAB91010.1 GAKP01004186 JAC54766.1 DS231890 EDS43796.1 GFDL01013565 JAV21480.1 AXCM01000185 UFQT01000883 SSX27832.1 GANO01000759 JAB59112.1 UFQT01000673 SSX26371.1 ATLV01014583 KE524975 KFB39098.1 GBXI01014177 JAD00115.1 GDAI01002514 JAI15089.1 GDHF01021417 JAI30897.1 PYGN01001714 PSN33237.1 LBMM01000086 KMR05021.1 QOIP01000007 RLU20092.1 EZ424132 ADD20408.1 CCAG010003788 GL888498 EGI60043.1 ADTU01012061 GL763883 EFZ19121.1 KQ981744 KYN36332.1 KQ434822 KZC07164.1 KQ982650 KYQ52918.1 KQ977012 KYN06344.1 KQ435818 KOX72538.1 NEVH01018389 PNF23393.1 KQ980278 KYN16966.1 GL442298 EFN63620.1 KK853272 KDR09129.1 KQ414661 KOC65591.1 GEZM01054792 JAV73396.1 JRES01000487 KNC30867.1 GL451412 EFN79396.1 KZ288242 PBC31254.1 AJVK01000607 AAZX01005683 CH379069 EAL31018.1 CH902618 EDV39168.1 CH940647 EDW70981.1 OUUW01000002 SPP76161.1 NNAY01001480 OXU23820.1 CH933809 EDW19359.1 GBYB01000082 GBYB01000409 JAG69849.1 JAG70176.1 CH963847 EDW73070.1 APGK01052181 KB741211 ENN72736.1 KB632263 ERL90872.1 CH480826 EDW44425.1 CH479188 EDW40485.1 CM000159 CH891688 EDW94965.1 EDW99634.1 CH916366 EDV96649.1 CM000363 CM002912 EDX11450.1 KMZ01101.1 KQ971351 EFA06469.1 CH954178 EDV52717.1 AF074334 AE014296 AAC26868.1 AAF51784.1 BT089027 ACU00260.1 BT023917 ABB36421.1 AF024618 AAB82275.1 GFDF01003206 JAV10878.1 GEBQ01013454 JAT26523.1
PCG74229.1 KQ459589 KPI97656.1 JTDY01004984 KOB67509.1 GAIX01003380 JAA89180.1 RSAL01000020 RVE52576.1 AGBW02007996 OWR54334.1 GDQN01011169 JAT79885.1 KQ460367 KPJ15504.1 CH477997 EAT34507.1 EAT34510.1 AAAB01008964 EAA12839.3 APCN01000774 JXUM01097524 KQ564351 KXJ72389.1 GGFJ01007226 MBW56367.1 GGFJ01007225 MBW56366.1 ADMH02001866 ETN60786.1 AXCN02000840 GAMC01015547 GAMC01015546 GAMC01015545 JAB91010.1 GAKP01004186 JAC54766.1 DS231890 EDS43796.1 GFDL01013565 JAV21480.1 AXCM01000185 UFQT01000883 SSX27832.1 GANO01000759 JAB59112.1 UFQT01000673 SSX26371.1 ATLV01014583 KE524975 KFB39098.1 GBXI01014177 JAD00115.1 GDAI01002514 JAI15089.1 GDHF01021417 JAI30897.1 PYGN01001714 PSN33237.1 LBMM01000086 KMR05021.1 QOIP01000007 RLU20092.1 EZ424132 ADD20408.1 CCAG010003788 GL888498 EGI60043.1 ADTU01012061 GL763883 EFZ19121.1 KQ981744 KYN36332.1 KQ434822 KZC07164.1 KQ982650 KYQ52918.1 KQ977012 KYN06344.1 KQ435818 KOX72538.1 NEVH01018389 PNF23393.1 KQ980278 KYN16966.1 GL442298 EFN63620.1 KK853272 KDR09129.1 KQ414661 KOC65591.1 GEZM01054792 JAV73396.1 JRES01000487 KNC30867.1 GL451412 EFN79396.1 KZ288242 PBC31254.1 AJVK01000607 AAZX01005683 CH379069 EAL31018.1 CH902618 EDV39168.1 CH940647 EDW70981.1 OUUW01000002 SPP76161.1 NNAY01001480 OXU23820.1 CH933809 EDW19359.1 GBYB01000082 GBYB01000409 JAG69849.1 JAG70176.1 CH963847 EDW73070.1 APGK01052181 KB741211 ENN72736.1 KB632263 ERL90872.1 CH480826 EDW44425.1 CH479188 EDW40485.1 CM000159 CH891688 EDW94965.1 EDW99634.1 CH916366 EDV96649.1 CM000363 CM002912 EDX11450.1 KMZ01101.1 KQ971351 EFA06469.1 CH954178 EDV52717.1 AF074334 AE014296 AAC26868.1 AAF51784.1 BT089027 ACU00260.1 BT023917 ABB36421.1 AF024618 AAB82275.1 GFDF01003206 JAV10878.1 GEBQ01013454 JAT26523.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037510
UP000283053
UP000007151
+ More
UP000053240 UP000008820 UP000075903 UP000075882 UP000007062 UP000075840 UP000069940 UP000249989 UP000075902 UP000076407 UP000075881 UP000000673 UP000075901 UP000075920 UP000076408 UP000075900 UP000075884 UP000069272 UP000075885 UP000075886 UP000075880 UP000002320 UP000075883 UP000030765 UP000245037 UP000095301 UP000036403 UP000279307 UP000092444 UP000092445 UP000007755 UP000005205 UP000078541 UP000076502 UP000075809 UP000078542 UP000053105 UP000235965 UP000078492 UP000000311 UP000005203 UP000027135 UP000078200 UP000053825 UP000037069 UP000008237 UP000092443 UP000242457 UP000192223 UP000092462 UP000002358 UP000001819 UP000007801 UP000008792 UP000268350 UP000215335 UP000192221 UP000009192 UP000007798 UP000019118 UP000030742 UP000001292 UP000008744 UP000002282 UP000001070 UP000000304 UP000007266 UP000008711 UP000000803 UP000091820 UP000079169
UP000053240 UP000008820 UP000075903 UP000075882 UP000007062 UP000075840 UP000069940 UP000249989 UP000075902 UP000076407 UP000075881 UP000000673 UP000075901 UP000075920 UP000076408 UP000075900 UP000075884 UP000069272 UP000075885 UP000075886 UP000075880 UP000002320 UP000075883 UP000030765 UP000245037 UP000095301 UP000036403 UP000279307 UP000092444 UP000092445 UP000007755 UP000005205 UP000078541 UP000076502 UP000075809 UP000078542 UP000053105 UP000235965 UP000078492 UP000000311 UP000005203 UP000027135 UP000078200 UP000053825 UP000037069 UP000008237 UP000092443 UP000242457 UP000192223 UP000092462 UP000002358 UP000001819 UP000007801 UP000008792 UP000268350 UP000215335 UP000192221 UP000009192 UP000007798 UP000019118 UP000030742 UP000001292 UP000008744 UP000002282 UP000001070 UP000000304 UP000007266 UP000008711 UP000000803 UP000091820 UP000079169
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
H9JGM4
A0A2H1WI10
A0A2W1BQZ3
A0A2A4JQZ1
A0A194PWA2
A0A0L7KWS8
+ More
S4PMU2 A0A3S2PIR6 A0A212FKR7 A0A1E1VYT2 A0A194RDG2 Q16JQ2 A0A182UXN1 A0A182L0E9 Q7Q4I7 A0A182IC32 A0A182GYI0 A0A182UFH5 A0A2M4BTG6 A0A2M4BTE8 A0A182XFU7 A0A182K6S6 W5J9W4 A0A182SFC9 A0A182WHC3 A0A182Y9Y1 A0A182RG34 A0A182NET9 A0A182F5K1 A0A182PAP1 A0A182QFH8 A0A182IJV9 W8AQE1 A0A034WGT5 B0WCS0 A0A1Q3F1P6 A0A182M243 A0A336MBU1 U5EZI0 A0A336MAL8 A0A084VMA4 A0A0A1WNN0 A0A0K8TL95 A0A0K8UW44 A0A2P8XMK8 A0A1I8MZY5 A0A0J7LA15 A0A3L8DIR2 D3TRT0 A0A1B0G766 A0A1A9ZXB0 F4X0Q1 A0A158NCY3 E9IJX2 A0A195F817 A0A154P750 A0A151WYF4 A0A195D1R5 A0A0N0BEY2 A0A2J7Q483 A0A151J3D1 E2ASF4 A0A087ZN02 A0A067QJI2 A0A1A9UQ25 A0A0L7R4B0 A0A1Y1LP72 A0A0L0CF97 E2BY23 A0A1A9YB41 A0A2A3EIF7 A0A1W4WY47 A0A1B0DB14 K7IUX2 Q2M0B7 B3MAT4 B4LCT9 A0A3B0JTX4 A0A232EZX0 A0A1W4USX4 B4KUV3 A0A0C9QHN8 B4MM61 N6TWG3 U4UCW3 B4IAW2 B4GRV0 B4ITC1 B4IZ94 B4QKN5 D6WRD7 B3NJ00 O76513 C6SV35 Q32KG2 O17144 A0A1A9WQR9 A0A1L8DWS4 A0A1B6LS70 A0A1S4ESS3
S4PMU2 A0A3S2PIR6 A0A212FKR7 A0A1E1VYT2 A0A194RDG2 Q16JQ2 A0A182UXN1 A0A182L0E9 Q7Q4I7 A0A182IC32 A0A182GYI0 A0A182UFH5 A0A2M4BTG6 A0A2M4BTE8 A0A182XFU7 A0A182K6S6 W5J9W4 A0A182SFC9 A0A182WHC3 A0A182Y9Y1 A0A182RG34 A0A182NET9 A0A182F5K1 A0A182PAP1 A0A182QFH8 A0A182IJV9 W8AQE1 A0A034WGT5 B0WCS0 A0A1Q3F1P6 A0A182M243 A0A336MBU1 U5EZI0 A0A336MAL8 A0A084VMA4 A0A0A1WNN0 A0A0K8TL95 A0A0K8UW44 A0A2P8XMK8 A0A1I8MZY5 A0A0J7LA15 A0A3L8DIR2 D3TRT0 A0A1B0G766 A0A1A9ZXB0 F4X0Q1 A0A158NCY3 E9IJX2 A0A195F817 A0A154P750 A0A151WYF4 A0A195D1R5 A0A0N0BEY2 A0A2J7Q483 A0A151J3D1 E2ASF4 A0A087ZN02 A0A067QJI2 A0A1A9UQ25 A0A0L7R4B0 A0A1Y1LP72 A0A0L0CF97 E2BY23 A0A1A9YB41 A0A2A3EIF7 A0A1W4WY47 A0A1B0DB14 K7IUX2 Q2M0B7 B3MAT4 B4LCT9 A0A3B0JTX4 A0A232EZX0 A0A1W4USX4 B4KUV3 A0A0C9QHN8 B4MM61 N6TWG3 U4UCW3 B4IAW2 B4GRV0 B4ITC1 B4IZ94 B4QKN5 D6WRD7 B3NJ00 O76513 C6SV35 Q32KG2 O17144 A0A1A9WQR9 A0A1L8DWS4 A0A1B6LS70 A0A1S4ESS3
PDB
1JKW
E-value=8.90049e-64,
Score=617
Ontologies
PATHWAY
GO
Topology
Subcellular location
Secreted
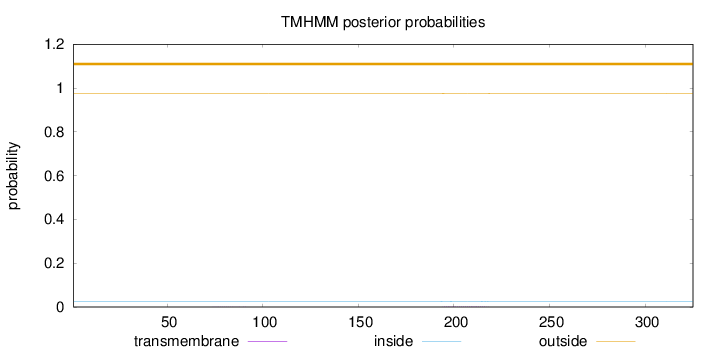
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04695
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02468
outside
1 - 325
Population Genetic Test Statistics
Pi
240.866549
Theta
220.913634
Tajima's D
0.110126
CLR
6.886665
CSRT
0.401829908504575
Interpretation
Uncertain
