Gene
KWMTBOMO04159
Pre Gene Modal
BGIBMGA008674
Annotation
unc-50-like_protein_[Bombyx_mori]
Full name
Protein unc-50 homolog
+ More
Mothers against decapentaplegic homolog
Mothers against decapentaplegic homolog
Alternative Name
Uncoordinated-like protein
SMAD family member
SMAD family member
Location in the cell
PlasmaMembrane Reliability : 4.893
Sequence
CDS
ATGAAGTATTCGACGTCGCCTACTCCAAATCTTCATAACTATCCTAGGAGCACTTCACCCCTGCCAGCCCCGGCTAATTATCAGCCAACAACAGCTAGCGCTTCAGTAAAACGCTACAAATACCTCAAGAGACTGTTCAAATTTAATCAAATGGACTTCGAATTCGCCGCTTGGCAAATGGTTTACTTGTTCATTGCCCCTCAGAAAGTATTCCGAAACTTTAACTATAGGAAGCATACAAAATCACAGTTTGCGAGAGACGACCCTGCATTCTTAGTGTTGCTCAGCATCTGGTTATTTTTGTCGTCAATATGTTTCGGATTGGCACTAGACCTGACTGTGGGCAAAGTTGCATTATTCATGCTGTTTGTGGTGTTCGTTGATTTCATCGGAGCAGGAATACTGGTCTCCACATTATTTTGGTACCTGAGCAACAAGCACCTGCGCCGCGACCCCGAGGGTCCGGACGTGGAGTGGGGCTACGCCTTCGACGTGCACATCAACGCGTTTTTCCCGCCGCTCTCGCTGCTGCACTGCTTCCAAATCGTACTATTCAACAGTATATTGTCTCAAGCGGGCTTCGTCTCGTGTCTGGTCTCAAACACCTTCTGGCTCGCCTCGATCATATACTACATGTACATTTCCTTCCTGGGATACAGCAACCTGCCGTTCCTGCAAAACACGAGGGTGTTCCTGCTGCCGCTGCCCGTGCTATTCGTGGTCTACCTGGCCACCCTGCTGGCCGGCTGGAACCTCAGCGAGCTGCTCATGTACTTCTATAGGTACAGGGTGTTGTAG
Protein
MKYSTSPTPNLHNYPRSTSPLPAPANYQPTTASASVKRYKYLKRLFKFNQMDFEFAAWQMVYLFIAPQKVFRNFNYRKHTKSQFARDDPAFLVLLSIWLFLSSICFGLALDLTVGKVALFMLFVVFVDFIGAGILVSTLFWYLSNKHLRRDPEGPDVEWGYAFDVHINAFFPPLSLLHCFQIVLFNSILSQAGFVSCLVSNTFWLASIIYYMYISFLGYSNLPFLQNTRVFLLPLPVLFVVYLATLLAGWNLSELLMYFYRYRVL
Summary
Description
May be required for cell surface expression of acetylcholine receptors.
May be involved in cell surface expression of neuronal nicotinic receptors. Binds RNA (By similarity).
May be involved in cell surface expression of neuronal nicotinic receptors. Binds RNA (By similarity).
Similarity
Belongs to the unc-50 family.
Belongs to the dwarfin/SMAD family.
Belongs to the dwarfin/SMAD family.
Keywords
Complete proteome
Golgi apparatus
Membrane
Protein transport
Reference proteome
Transmembrane
Transmembrane helix
Transport
Acetylation
Alternative splicing
Nucleus
Phosphoprotein
RNA-binding
Feature
chain Protein unc-50 homolog
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
Q2F641
A0A2H1W6L8
A0A2A4JQD8
A0A2W1BXP4
A0A194PWB2
I4DL67
+ More
S4PME3 A0A023EM42 Q16HU6 B0W696 A0A1Q3FAF7 A0A182WMW6 A0A182LTQ7 A0A1I8NZS7 A0A1I8MHI8 A0A182QRM1 A0A0K8VNC1 A0A182TH78 A0A182JHJ6 A0A182VMP3 A0A182LG82 Q7PR08 W5JD79 B4LZN6 A0A2M4AK20 A0A2M3ZKY0 A0A0M4EST5 A0A182N549 D6W8C1 A0A0K8TMX9 Q294K8 B4GMY8 A0A2M4BWL6 B3LXG3 B4K7J3 A0A067R501 D0QWL5 A0A0L0CKJ0 A0A2M4BWT7 A0A0T6AWA0 A0A1W4XFD0 E0VHN0 A0A3B0JUI0 T1E8A1 A0A1B6DIF3 A0A210Q5J0 A0A1B6IS62 U5EV70 A0A1B6ERW1 A0A1W4W446 A0A1B0AA20 D3TNE3 B4NB00 Q9VHN5 A0A1A9YEQ4 E1ZYA3 B4QXZ7 B4PT86 B4HL34 B3P4U8 E9J004 A0A1A9VDZ5 A0A1B0BEV1 A0A1B6KPT4 A0A026W215 A0A0L8H648 A0A0V0G6V6 A0A182HTV9 B4JI03 A0A182JNI3 A0A088A0U2 A0A0M8ZXW0 A0A2R7WSH3 G1P116 A0A0L7QQB8 A0A2A3EMM7 A0A182XHI7 A0A182H6G8 A0A154PBX5 A0A151J4C6 B5DEV8 A0A0P6JC01 A0A195BTB8 A0A158NB45 R7UNM3 A0A195EW25 G5BMT5 A0A091KPV7 L8HP02 Q3ZBG6 W5KZ39 A0A2P2I278 F4WHB2 I3LLY5 A0A3L7INC0 A0A1U7QBG0 W5PVK2 A0A0A9YVQ2 A0A337SQ93
S4PME3 A0A023EM42 Q16HU6 B0W696 A0A1Q3FAF7 A0A182WMW6 A0A182LTQ7 A0A1I8NZS7 A0A1I8MHI8 A0A182QRM1 A0A0K8VNC1 A0A182TH78 A0A182JHJ6 A0A182VMP3 A0A182LG82 Q7PR08 W5JD79 B4LZN6 A0A2M4AK20 A0A2M3ZKY0 A0A0M4EST5 A0A182N549 D6W8C1 A0A0K8TMX9 Q294K8 B4GMY8 A0A2M4BWL6 B3LXG3 B4K7J3 A0A067R501 D0QWL5 A0A0L0CKJ0 A0A2M4BWT7 A0A0T6AWA0 A0A1W4XFD0 E0VHN0 A0A3B0JUI0 T1E8A1 A0A1B6DIF3 A0A210Q5J0 A0A1B6IS62 U5EV70 A0A1B6ERW1 A0A1W4W446 A0A1B0AA20 D3TNE3 B4NB00 Q9VHN5 A0A1A9YEQ4 E1ZYA3 B4QXZ7 B4PT86 B4HL34 B3P4U8 E9J004 A0A1A9VDZ5 A0A1B0BEV1 A0A1B6KPT4 A0A026W215 A0A0L8H648 A0A0V0G6V6 A0A182HTV9 B4JI03 A0A182JNI3 A0A088A0U2 A0A0M8ZXW0 A0A2R7WSH3 G1P116 A0A0L7QQB8 A0A2A3EMM7 A0A182XHI7 A0A182H6G8 A0A154PBX5 A0A151J4C6 B5DEV8 A0A0P6JC01 A0A195BTB8 A0A158NB45 R7UNM3 A0A195EW25 G5BMT5 A0A091KPV7 L8HP02 Q3ZBG6 W5KZ39 A0A2P2I278 F4WHB2 I3LLY5 A0A3L7INC0 A0A1U7QBG0 W5PVK2 A0A0A9YVQ2 A0A337SQ93
Pubmed
19121390
28756777
26354079
22651552
23622113
24945155
+ More
26483478 17510324 25315136 20966253 12364791 14747013 17210077 20920257 23761445 17994087 18362917 19820115 26369729 15632085 24845553 19859648 26108605 20566863 28812685 20353571 10731132 12537572 12537569 10980252 20798317 17550304 21282665 24508170 30249741 21993624 20431018 21347285 23254933 21993625 22751099 16305752 25329095 21719571 29704459 20809919 25401762 17975172
26483478 17510324 25315136 20966253 12364791 14747013 17210077 20920257 23761445 17994087 18362917 19820115 26369729 15632085 24845553 19859648 26108605 20566863 28812685 20353571 10731132 12537572 12537569 10980252 20798317 17550304 21282665 24508170 30249741 21993624 20431018 21347285 23254933 21993625 22751099 16305752 25329095 21719571 29704459 20809919 25401762 17975172
EMBL
BABH01034372
DQ311231
ABD36176.1
ODYU01006674
SOQ48730.1
NWSH01000785
+ More
PCG74211.1 KZ149929 PZC77456.1 KQ459589 KPI97666.1 AK402035 BAM18657.1 GAIX01003600 JAA88960.1 JXUM01090470 GAPW01003241 KQ563841 JAC10357.1 KXJ73185.1 CH478140 EAT33828.1 DS231847 EDS36443.1 GFDL01010494 JAV24551.1 AXCM01004558 AXCN02000174 GDHF01011935 JAI40379.1 AAAB01008859 EAA08030.4 ADMH02001856 ETN60850.1 CH940650 EDW68205.1 GGFK01007814 MBW41135.1 GGFM01008438 MBW29189.1 CP012526 ALC47860.1 KQ971307 EFA10930.1 GDAI01001899 JAI15704.1 CM000070 EAL28959.1 CH479185 EDW38212.1 GGFJ01008334 MBW57475.1 CH902617 EDV42807.1 CH933806 EDW15337.1 KK852747 KDR17270.1 FJ821033 ACN94714.1 JRES01000266 KNC32786.1 GGFJ01008333 MBW57474.1 LJIG01022715 KRT79086.1 DS235171 EEB12916.1 OUUW01000008 SPP84052.1 GAMD01002357 JAA99233.1 GEDC01030973 GEDC01011866 JAS06325.1 JAS25432.1 NEDP02004947 OWF43985.1 GECU01017983 JAS89723.1 GANO01002012 JAB57859.1 GECZ01029058 JAS40711.1 EZ422945 ADD19221.1 CH964232 EDW80964.1 AE014297 AY113422 GL435204 EFN73802.1 CM000364 EDX13734.1 CM000160 EDW96547.1 CH480815 EDW42998.1 CH954181 EDV49892.1 GL767291 EFZ13812.1 JXJN01013026 GEBQ01026525 JAT13452.1 KK107485 QOIP01000006 EZA50083.1 RLU21932.1 KQ419106 KOF84574.1 GECL01002382 JAP03742.1 APCN01000586 CH916369 EDV92911.1 KJ939605 AJE70276.1 KQ435830 KOX71799.1 KK855421 PTY22503.1 AAPE02034870 KQ414797 KOC60691.1 KZ288208 PBC33015.1 JXUM01027095 JXUM01027096 JXUM01027097 KQ560779 KXJ80923.1 KQ434869 KZC09419.1 KQ980151 KYN17491.1 AAMC01074123 AAMC01074124 BC168819 BC169164 AAI68819.1 GEBF01005543 JAN98089.1 KQ976417 KYM89681.1 ADTU01010875 AMQN01006852 AMQN01006853 KB299334 ELU08114.1 KQ981953 KYN32351.1 JH171109 EHB10596.1 KK751870 KFP42052.1 JH883698 ELR46035.1 BT021031 BC103305 IACF01002437 LAB68091.1 GL888158 EGI66389.1 AEMK02000020 RAZU01000007 RLQ79617.1 AMGL01078775 GBHO01007305 JAG36299.1 AANG04002548
PCG74211.1 KZ149929 PZC77456.1 KQ459589 KPI97666.1 AK402035 BAM18657.1 GAIX01003600 JAA88960.1 JXUM01090470 GAPW01003241 KQ563841 JAC10357.1 KXJ73185.1 CH478140 EAT33828.1 DS231847 EDS36443.1 GFDL01010494 JAV24551.1 AXCM01004558 AXCN02000174 GDHF01011935 JAI40379.1 AAAB01008859 EAA08030.4 ADMH02001856 ETN60850.1 CH940650 EDW68205.1 GGFK01007814 MBW41135.1 GGFM01008438 MBW29189.1 CP012526 ALC47860.1 KQ971307 EFA10930.1 GDAI01001899 JAI15704.1 CM000070 EAL28959.1 CH479185 EDW38212.1 GGFJ01008334 MBW57475.1 CH902617 EDV42807.1 CH933806 EDW15337.1 KK852747 KDR17270.1 FJ821033 ACN94714.1 JRES01000266 KNC32786.1 GGFJ01008333 MBW57474.1 LJIG01022715 KRT79086.1 DS235171 EEB12916.1 OUUW01000008 SPP84052.1 GAMD01002357 JAA99233.1 GEDC01030973 GEDC01011866 JAS06325.1 JAS25432.1 NEDP02004947 OWF43985.1 GECU01017983 JAS89723.1 GANO01002012 JAB57859.1 GECZ01029058 JAS40711.1 EZ422945 ADD19221.1 CH964232 EDW80964.1 AE014297 AY113422 GL435204 EFN73802.1 CM000364 EDX13734.1 CM000160 EDW96547.1 CH480815 EDW42998.1 CH954181 EDV49892.1 GL767291 EFZ13812.1 JXJN01013026 GEBQ01026525 JAT13452.1 KK107485 QOIP01000006 EZA50083.1 RLU21932.1 KQ419106 KOF84574.1 GECL01002382 JAP03742.1 APCN01000586 CH916369 EDV92911.1 KJ939605 AJE70276.1 KQ435830 KOX71799.1 KK855421 PTY22503.1 AAPE02034870 KQ414797 KOC60691.1 KZ288208 PBC33015.1 JXUM01027095 JXUM01027096 JXUM01027097 KQ560779 KXJ80923.1 KQ434869 KZC09419.1 KQ980151 KYN17491.1 AAMC01074123 AAMC01074124 BC168819 BC169164 AAI68819.1 GEBF01005543 JAN98089.1 KQ976417 KYM89681.1 ADTU01010875 AMQN01006852 AMQN01006853 KB299334 ELU08114.1 KQ981953 KYN32351.1 JH171109 EHB10596.1 KK751870 KFP42052.1 JH883698 ELR46035.1 BT021031 BC103305 IACF01002437 LAB68091.1 GL888158 EGI66389.1 AEMK02000020 RAZU01000007 RLQ79617.1 AMGL01078775 GBHO01007305 JAG36299.1 AANG04002548
Proteomes
UP000005204
UP000218220
UP000053268
UP000069940
UP000249989
UP000008820
+ More
UP000002320 UP000075920 UP000075883 UP000095300 UP000095301 UP000075886 UP000075902 UP000075880 UP000075903 UP000075882 UP000007062 UP000000673 UP000008792 UP000092553 UP000075884 UP000007266 UP000001819 UP000008744 UP000007801 UP000009192 UP000027135 UP000037069 UP000192223 UP000009046 UP000268350 UP000242188 UP000192221 UP000092445 UP000007798 UP000000803 UP000092443 UP000000311 UP000000304 UP000002282 UP000001292 UP000008711 UP000078200 UP000092460 UP000053097 UP000279307 UP000053454 UP000075840 UP000001070 UP000075881 UP000005203 UP000053105 UP000001074 UP000053825 UP000242457 UP000076407 UP000076502 UP000078492 UP000008143 UP000078540 UP000005205 UP000014760 UP000078541 UP000006813 UP000009136 UP000018467 UP000007755 UP000008227 UP000273346 UP000189706 UP000002356 UP000011712
UP000002320 UP000075920 UP000075883 UP000095300 UP000095301 UP000075886 UP000075902 UP000075880 UP000075903 UP000075882 UP000007062 UP000000673 UP000008792 UP000092553 UP000075884 UP000007266 UP000001819 UP000008744 UP000007801 UP000009192 UP000027135 UP000037069 UP000192223 UP000009046 UP000268350 UP000242188 UP000192221 UP000092445 UP000007798 UP000000803 UP000092443 UP000000311 UP000000304 UP000002282 UP000001292 UP000008711 UP000078200 UP000092460 UP000053097 UP000279307 UP000053454 UP000075840 UP000001070 UP000075881 UP000005203 UP000053105 UP000001074 UP000053825 UP000242457 UP000076407 UP000076502 UP000078492 UP000008143 UP000078540 UP000005205 UP000014760 UP000078541 UP000006813 UP000009136 UP000018467 UP000007755 UP000008227 UP000273346 UP000189706 UP000002356 UP000011712
Interpro
Gene 3D
ProteinModelPortal
Q2F641
A0A2H1W6L8
A0A2A4JQD8
A0A2W1BXP4
A0A194PWB2
I4DL67
+ More
S4PME3 A0A023EM42 Q16HU6 B0W696 A0A1Q3FAF7 A0A182WMW6 A0A182LTQ7 A0A1I8NZS7 A0A1I8MHI8 A0A182QRM1 A0A0K8VNC1 A0A182TH78 A0A182JHJ6 A0A182VMP3 A0A182LG82 Q7PR08 W5JD79 B4LZN6 A0A2M4AK20 A0A2M3ZKY0 A0A0M4EST5 A0A182N549 D6W8C1 A0A0K8TMX9 Q294K8 B4GMY8 A0A2M4BWL6 B3LXG3 B4K7J3 A0A067R501 D0QWL5 A0A0L0CKJ0 A0A2M4BWT7 A0A0T6AWA0 A0A1W4XFD0 E0VHN0 A0A3B0JUI0 T1E8A1 A0A1B6DIF3 A0A210Q5J0 A0A1B6IS62 U5EV70 A0A1B6ERW1 A0A1W4W446 A0A1B0AA20 D3TNE3 B4NB00 Q9VHN5 A0A1A9YEQ4 E1ZYA3 B4QXZ7 B4PT86 B4HL34 B3P4U8 E9J004 A0A1A9VDZ5 A0A1B0BEV1 A0A1B6KPT4 A0A026W215 A0A0L8H648 A0A0V0G6V6 A0A182HTV9 B4JI03 A0A182JNI3 A0A088A0U2 A0A0M8ZXW0 A0A2R7WSH3 G1P116 A0A0L7QQB8 A0A2A3EMM7 A0A182XHI7 A0A182H6G8 A0A154PBX5 A0A151J4C6 B5DEV8 A0A0P6JC01 A0A195BTB8 A0A158NB45 R7UNM3 A0A195EW25 G5BMT5 A0A091KPV7 L8HP02 Q3ZBG6 W5KZ39 A0A2P2I278 F4WHB2 I3LLY5 A0A3L7INC0 A0A1U7QBG0 W5PVK2 A0A0A9YVQ2 A0A337SQ93
S4PME3 A0A023EM42 Q16HU6 B0W696 A0A1Q3FAF7 A0A182WMW6 A0A182LTQ7 A0A1I8NZS7 A0A1I8MHI8 A0A182QRM1 A0A0K8VNC1 A0A182TH78 A0A182JHJ6 A0A182VMP3 A0A182LG82 Q7PR08 W5JD79 B4LZN6 A0A2M4AK20 A0A2M3ZKY0 A0A0M4EST5 A0A182N549 D6W8C1 A0A0K8TMX9 Q294K8 B4GMY8 A0A2M4BWL6 B3LXG3 B4K7J3 A0A067R501 D0QWL5 A0A0L0CKJ0 A0A2M4BWT7 A0A0T6AWA0 A0A1W4XFD0 E0VHN0 A0A3B0JUI0 T1E8A1 A0A1B6DIF3 A0A210Q5J0 A0A1B6IS62 U5EV70 A0A1B6ERW1 A0A1W4W446 A0A1B0AA20 D3TNE3 B4NB00 Q9VHN5 A0A1A9YEQ4 E1ZYA3 B4QXZ7 B4PT86 B4HL34 B3P4U8 E9J004 A0A1A9VDZ5 A0A1B0BEV1 A0A1B6KPT4 A0A026W215 A0A0L8H648 A0A0V0G6V6 A0A182HTV9 B4JI03 A0A182JNI3 A0A088A0U2 A0A0M8ZXW0 A0A2R7WSH3 G1P116 A0A0L7QQB8 A0A2A3EMM7 A0A182XHI7 A0A182H6G8 A0A154PBX5 A0A151J4C6 B5DEV8 A0A0P6JC01 A0A195BTB8 A0A158NB45 R7UNM3 A0A195EW25 G5BMT5 A0A091KPV7 L8HP02 Q3ZBG6 W5KZ39 A0A2P2I278 F4WHB2 I3LLY5 A0A3L7INC0 A0A1U7QBG0 W5PVK2 A0A0A9YVQ2 A0A337SQ93
Ontologies
GO
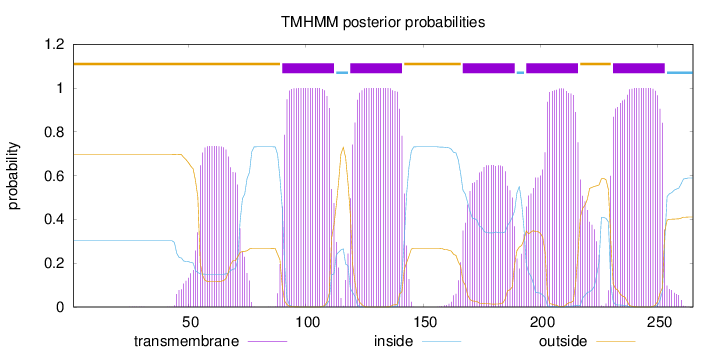
Topology
Subcellular location
Golgi apparatus membrane
Cytoplasm
Nucleus
Nucleus inner membrane
Cytoplasm
Nucleus
Nucleus inner membrane
Length:
265
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
118.66136
Exp number, first 60 AAs:
5.79137
Total prob of N-in:
0.30262
outside
1 - 89
TMhelix
90 - 112
inside
113 - 118
TMhelix
119 - 141
outside
142 - 166
TMhelix
167 - 189
inside
190 - 193
TMhelix
194 - 216
outside
217 - 230
TMhelix
231 - 253
inside
254 - 265
Population Genetic Test Statistics
Pi
54.291412
Theta
152.149784
Tajima's D
-1.587846
CLR
1282.739486
CSRT
0.0508474576271186
Interpretation
Uncertain
