Pre Gene Modal
BGIBMGA008718
Annotation
hypothetical_protein_KGM_20880_[Danaus_plexippus]
Full name
DNA-directed RNA polymerase III subunit RPC4
Alternative Name
DNA-directed RNA polymerase III subunit D
Location in the cell
Nuclear Reliability : 3.066
Sequence
CDS
ATGTCCGGTGATAAATCATCTAAACCGAATGGACTGGGTACAGATCCCATGCAGAGGCTCGCTTCATTAAAAGCACCCAGAGATCTTATGCTTGGAGGAGTCAAACCAAACAAAAAAGTCTTCACACCTAACTTGAACGTAGCTCGGAGTAAAAATAAAGGACAAGCAACAACAAGTGCCAAAGAAACAAGGAATGAAGAGAAGAACAGCAGGGATTCAAACAGGAACCGGAATTTTAACAGGAATCAGAGAAATGGGCCAACACTAATAAAATCTGCTGGTGTCTTCTCTGAAGGCATCGGTTCATCAGAGAGGCATTCTTCAAGTCGCTCGTACTACGGGCGGGACACAGAAAAAACTACAAACATTATGAAGAAGCCAGCTATCAGAGTAAAGGATTTCGTGAAGATCGACAAAGAACTGGAGGAGAAGAAGATAAATATGATTTTAAGAGGAGGCGATACTGACGAGCTGGAGTCCATCGACTTTAAGGACATCCTCGACAGGGACGCACCCATCAAACTACCCATGGATGATGGGAACTGGGCCAAGACTCATGCGAAACCGAAAGTCAAGGTTAAAGAAGAGATTGTTGTCAAGACAGAACCCCGGGAAGATGGCGACTGTGTTATAAGCACACCAGAACGAAAGCCAGACGTGAAGGAAGCGTTTGAAGATACCGACTTAGTGAACTTACTTAATAACGACAAATCAACATTAATTATGTTGCAGCTTCCCTCGAGTCTCCCGGGCAGAGGCGGCCGCGTCGAGGACGACGTTCCCCGGAGGAGGACCAAGAACGAGCCCGGCCCGTCCGCCGGGGGGGGCGCGGACCCCGGCGCGGCGGACGAGCAGGAGGAGGACAACAAGTGTCGCCTGACGGACCTCGAGGAGGGTAGGATCGGCAAGCTGCAGCTCCACAGGTCGGGGAGGGTCACCCTGGCTTTAGGAGAAACTGTATTTGAGGTATCGATGGGCACAAAAGCTGCGTTCCACCAAGAACTGGTTTCTGTAAGTACTGATGACGCGTCACGCTCAGCCAAGGTGACCTCCTTAGGCGCTCTCCGACATAAACTAAACGTGATCCCGGATTGGGAAACAATGTTTCGGGATATTCCTCTATAG
Protein
MSGDKSSKPNGLGTDPMQRLASLKAPRDLMLGGVKPNKKVFTPNLNVARSKNKGQATTSAKETRNEEKNSRDSNRNRNFNRNQRNGPTLIKSAGVFSEGIGSSERHSSSRSYYGRDTEKTTNIMKKPAIRVKDFVKIDKELEEKKINMILRGGDTDELESIDFKDILDRDAPIKLPMDDGNWAKTHAKPKVKVKEEIVVKTEPREDGDCVISTPERKPDVKEAFEDTDLVNLLNNDKSTLIMLQLPSSLPGRGGRVEDDVPRRRTKNEPGPSAGGGADPGAADEQEEDNKCRLTDLEEGRIGKLQLHRSGRVTLALGETVFEVSMGTKAAFHQELVSVSTDDASRSAKVTSLGALRHKLNVIPDWETMFRDIPL
Summary
Description
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs. Plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses. Acts as nuclear and cytosolic DNA sensor involved in innate immune response. Can sense non-self dsDNA that serves as template for transcription into dsRNA. The non-self RNA polymerase III transcripts induce type I interferon and NF- Kappa-B through the RIG-I pathway (By similarity).
Subunit
Component of the RNA polymerase III (Pol III) complex consisting of 17 subunits. Interacts with RPC5 (By similarity).
Similarity
Belongs to the eukaryotic RPC4/POLR3D RNA polymerase subunit family.
Keywords
Acetylation
Antiviral defense
Complete proteome
DNA-directed RNA polymerase
Immunity
Innate immunity
Isopeptide bond
Methylation
Nucleus
Phosphoprotein
Reference proteome
Transcription
Ubl conjugation
Feature
chain DNA-directed RNA polymerase III subunit RPC4
Uniprot
H9JGS1
A0A2A4JQ70
A0A2H1W6S1
A0A2A4JRI3
A0A194PY41
A0A212FPJ4
+ More
A0A1E1W2A5 A0A1E1WPD6 A0A1E1WQH3 A0A2W1BQT0 Q16XP6 A0A1Q3F2I1 B0WKN9 A0A084VRH5 A0A067QV31 A0A182JWD6 A0A182V4W7 A0A182NSF5 Q5TSX9 A0A182PP91 A0A2M3ZBA8 A0A182KZL2 A0A182XFD4 A0A182RZL3 A0A182W3E6 A0A182HYM7 A0A182Y8T8 A0A182JDW8 A0A2M4ATE8 A0A2M4AUN6 A0A182SJA4 W5JDB9 A0A2M4BS17 A0A182FQV6 A0A088AHL1 A0A2A3EBK7 A0A1A9X101 A0A151XCG1 A0A182Q487 A0A1B6KUG5 A0A0V0G870 A0A1B6CWU7 A0A0K8V9D8 A0A0K8UI44 A0A151NBL1 A0A1A9YCK7 A0A1B0AQM1 A0A1U7RTF6 A0A2K6LHE7 A0A2K6QE16 A0A1U7SE94 A0A2K5KFE9 A0A1U7S492 A0A2K5YG24 A0A096NU72 A0A2K5L6N0 G7PCV0 A0A2K6D862 G7N0N3 F7E6W4 K7FF32 L5L739 Q5E9Z7 K7FF40 A0A2U3W2Q6 A0A0D9RVE8 F1MTH4 A0A0L0C8C4 A0A1S3A4C7 A0A3B4X8E0 A0A034VCN0 A0A3Q1JRD4 A0A2K6TA70 A0A034VDA6 A0A2K5PLN3 A0A3Q7P6Y6 W5PKZ9 A0A2D0QX22 A0A1V4KHM4 A0A1A8Q6T2 A0A1A8LBM2 A0A1A8FZZ0 A0A2D0QX25 A0A1V4KHP6 A0A1A8RXU9 Q58D98
A0A1E1W2A5 A0A1E1WPD6 A0A1E1WQH3 A0A2W1BQT0 Q16XP6 A0A1Q3F2I1 B0WKN9 A0A084VRH5 A0A067QV31 A0A182JWD6 A0A182V4W7 A0A182NSF5 Q5TSX9 A0A182PP91 A0A2M3ZBA8 A0A182KZL2 A0A182XFD4 A0A182RZL3 A0A182W3E6 A0A182HYM7 A0A182Y8T8 A0A182JDW8 A0A2M4ATE8 A0A2M4AUN6 A0A182SJA4 W5JDB9 A0A2M4BS17 A0A182FQV6 A0A088AHL1 A0A2A3EBK7 A0A1A9X101 A0A151XCG1 A0A182Q487 A0A1B6KUG5 A0A0V0G870 A0A1B6CWU7 A0A0K8V9D8 A0A0K8UI44 A0A151NBL1 A0A1A9YCK7 A0A1B0AQM1 A0A1U7RTF6 A0A2K6LHE7 A0A2K6QE16 A0A1U7SE94 A0A2K5KFE9 A0A1U7S492 A0A2K5YG24 A0A096NU72 A0A2K5L6N0 G7PCV0 A0A2K6D862 G7N0N3 F7E6W4 K7FF32 L5L739 Q5E9Z7 K7FF40 A0A2U3W2Q6 A0A0D9RVE8 F1MTH4 A0A0L0C8C4 A0A1S3A4C7 A0A3B4X8E0 A0A034VCN0 A0A3Q1JRD4 A0A2K6TA70 A0A034VDA6 A0A2K5PLN3 A0A3Q7P6Y6 W5PKZ9 A0A2D0QX22 A0A1V4KHM4 A0A1A8Q6T2 A0A1A8LBM2 A0A1A8FZZ0 A0A2D0QX25 A0A1V4KHP6 A0A1A8RXU9 Q58D98
Pubmed
EMBL
BABH01034370
NWSH01000785
PCG74215.1
ODYU01006674
SOQ48733.1
PCG74214.1
+ More
KQ459589 KPI97669.1 AGBW02002137 OWR55664.1 GDQN01009934 JAT81120.1 GDQN01002202 JAT88852.1 GDQN01001983 JAT89071.1 KZ149929 PZC77459.1 CH477534 EAT39396.1 GFDL01013308 JAV21737.1 DS231974 EDS29970.1 ATLV01015669 KE525026 KFB40569.1 KK852982 KDR12904.1 AAAB01008898 EAL40548.4 GGFM01005041 MBW25792.1 APCN01005389 GGFK01010729 MBW44050.1 GGFK01010987 MBW44308.1 ADMH02001863 ETN60809.1 GGFJ01006708 MBW55849.1 KZ288310 PBC28566.1 KQ982314 KYQ58045.1 AXCN02000237 GEBQ01024892 JAT15085.1 GECL01002019 JAP04105.1 GEDC01019326 GEDC01004884 JAS17972.1 JAS32414.1 GDHF01017134 JAI35180.1 GDHF01025965 JAI26349.1 AKHW03003565 KYO34214.1 JXJN01001971 AHZZ02033108 AQIA01066973 CM001283 EHH64036.1 JU336032 JU476897 JV048007 CM001260 AFE79785.1 AFH33701.1 AFI38078.1 EHH28335.1 JSUE03040998 AGCU01168936 AGCU01168937 KB030270 ELK19250.1 BT020772 BC118212 AQIB01116627 JRES01000760 KNC28521.1 GAKP01019407 JAC39545.1 GAKP01019409 JAC39543.1 AMGL01048564 LSYS01003090 OPJ83949.1 HAEI01004473 SBR88957.1 HAEF01004630 HAEG01013787 SBR42012.1 HAEB01017814 HAEC01008101 SBQ64341.1 OPJ83948.1 HAEH01020275 SBS10537.1 BT021699 AAX46546.1
KQ459589 KPI97669.1 AGBW02002137 OWR55664.1 GDQN01009934 JAT81120.1 GDQN01002202 JAT88852.1 GDQN01001983 JAT89071.1 KZ149929 PZC77459.1 CH477534 EAT39396.1 GFDL01013308 JAV21737.1 DS231974 EDS29970.1 ATLV01015669 KE525026 KFB40569.1 KK852982 KDR12904.1 AAAB01008898 EAL40548.4 GGFM01005041 MBW25792.1 APCN01005389 GGFK01010729 MBW44050.1 GGFK01010987 MBW44308.1 ADMH02001863 ETN60809.1 GGFJ01006708 MBW55849.1 KZ288310 PBC28566.1 KQ982314 KYQ58045.1 AXCN02000237 GEBQ01024892 JAT15085.1 GECL01002019 JAP04105.1 GEDC01019326 GEDC01004884 JAS17972.1 JAS32414.1 GDHF01017134 JAI35180.1 GDHF01025965 JAI26349.1 AKHW03003565 KYO34214.1 JXJN01001971 AHZZ02033108 AQIA01066973 CM001283 EHH64036.1 JU336032 JU476897 JV048007 CM001260 AFE79785.1 AFH33701.1 AFI38078.1 EHH28335.1 JSUE03040998 AGCU01168936 AGCU01168937 KB030270 ELK19250.1 BT020772 BC118212 AQIB01116627 JRES01000760 KNC28521.1 GAKP01019407 JAC39545.1 GAKP01019409 JAC39543.1 AMGL01048564 LSYS01003090 OPJ83949.1 HAEI01004473 SBR88957.1 HAEF01004630 HAEG01013787 SBR42012.1 HAEB01017814 HAEC01008101 SBQ64341.1 OPJ83948.1 HAEH01020275 SBS10537.1 BT021699 AAX46546.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000008820
UP000002320
+ More
UP000030765 UP000027135 UP000075881 UP000075903 UP000075884 UP000007062 UP000075885 UP000075882 UP000076407 UP000075900 UP000075920 UP000075840 UP000076408 UP000075880 UP000075901 UP000000673 UP000069272 UP000005203 UP000242457 UP000091820 UP000075809 UP000075886 UP000050525 UP000092443 UP000092460 UP000189705 UP000233180 UP000233200 UP000233080 UP000233140 UP000028761 UP000233060 UP000009130 UP000233100 UP000233120 UP000006718 UP000007267 UP000010552 UP000009136 UP000245340 UP000029965 UP000037069 UP000079721 UP000261360 UP000265040 UP000233220 UP000233040 UP000286641 UP000002356 UP000221080 UP000190648
UP000030765 UP000027135 UP000075881 UP000075903 UP000075884 UP000007062 UP000075885 UP000075882 UP000076407 UP000075900 UP000075920 UP000075840 UP000076408 UP000075880 UP000075901 UP000000673 UP000069272 UP000005203 UP000242457 UP000091820 UP000075809 UP000075886 UP000050525 UP000092443 UP000092460 UP000189705 UP000233180 UP000233200 UP000233080 UP000233140 UP000028761 UP000233060 UP000009130 UP000233100 UP000233120 UP000006718 UP000007267 UP000010552 UP000009136 UP000245340 UP000029965 UP000037069 UP000079721 UP000261360 UP000265040 UP000233220 UP000233040 UP000286641 UP000002356 UP000221080 UP000190648
Pfam
PF05132 RNA_pol_Rpc4
Interpro
IPR007811
RPC4
ProteinModelPortal
H9JGS1
A0A2A4JQ70
A0A2H1W6S1
A0A2A4JRI3
A0A194PY41
A0A212FPJ4
+ More
A0A1E1W2A5 A0A1E1WPD6 A0A1E1WQH3 A0A2W1BQT0 Q16XP6 A0A1Q3F2I1 B0WKN9 A0A084VRH5 A0A067QV31 A0A182JWD6 A0A182V4W7 A0A182NSF5 Q5TSX9 A0A182PP91 A0A2M3ZBA8 A0A182KZL2 A0A182XFD4 A0A182RZL3 A0A182W3E6 A0A182HYM7 A0A182Y8T8 A0A182JDW8 A0A2M4ATE8 A0A2M4AUN6 A0A182SJA4 W5JDB9 A0A2M4BS17 A0A182FQV6 A0A088AHL1 A0A2A3EBK7 A0A1A9X101 A0A151XCG1 A0A182Q487 A0A1B6KUG5 A0A0V0G870 A0A1B6CWU7 A0A0K8V9D8 A0A0K8UI44 A0A151NBL1 A0A1A9YCK7 A0A1B0AQM1 A0A1U7RTF6 A0A2K6LHE7 A0A2K6QE16 A0A1U7SE94 A0A2K5KFE9 A0A1U7S492 A0A2K5YG24 A0A096NU72 A0A2K5L6N0 G7PCV0 A0A2K6D862 G7N0N3 F7E6W4 K7FF32 L5L739 Q5E9Z7 K7FF40 A0A2U3W2Q6 A0A0D9RVE8 F1MTH4 A0A0L0C8C4 A0A1S3A4C7 A0A3B4X8E0 A0A034VCN0 A0A3Q1JRD4 A0A2K6TA70 A0A034VDA6 A0A2K5PLN3 A0A3Q7P6Y6 W5PKZ9 A0A2D0QX22 A0A1V4KHM4 A0A1A8Q6T2 A0A1A8LBM2 A0A1A8FZZ0 A0A2D0QX25 A0A1V4KHP6 A0A1A8RXU9 Q58D98
A0A1E1W2A5 A0A1E1WPD6 A0A1E1WQH3 A0A2W1BQT0 Q16XP6 A0A1Q3F2I1 B0WKN9 A0A084VRH5 A0A067QV31 A0A182JWD6 A0A182V4W7 A0A182NSF5 Q5TSX9 A0A182PP91 A0A2M3ZBA8 A0A182KZL2 A0A182XFD4 A0A182RZL3 A0A182W3E6 A0A182HYM7 A0A182Y8T8 A0A182JDW8 A0A2M4ATE8 A0A2M4AUN6 A0A182SJA4 W5JDB9 A0A2M4BS17 A0A182FQV6 A0A088AHL1 A0A2A3EBK7 A0A1A9X101 A0A151XCG1 A0A182Q487 A0A1B6KUG5 A0A0V0G870 A0A1B6CWU7 A0A0K8V9D8 A0A0K8UI44 A0A151NBL1 A0A1A9YCK7 A0A1B0AQM1 A0A1U7RTF6 A0A2K6LHE7 A0A2K6QE16 A0A1U7SE94 A0A2K5KFE9 A0A1U7S492 A0A2K5YG24 A0A096NU72 A0A2K5L6N0 G7PCV0 A0A2K6D862 G7N0N3 F7E6W4 K7FF32 L5L739 Q5E9Z7 K7FF40 A0A2U3W2Q6 A0A0D9RVE8 F1MTH4 A0A0L0C8C4 A0A1S3A4C7 A0A3B4X8E0 A0A034VCN0 A0A3Q1JRD4 A0A2K6TA70 A0A034VDA6 A0A2K5PLN3 A0A3Q7P6Y6 W5PKZ9 A0A2D0QX22 A0A1V4KHM4 A0A1A8Q6T2 A0A1A8LBM2 A0A1A8FZZ0 A0A2D0QX25 A0A1V4KHP6 A0A1A8RXU9 Q58D98
PDB
6F44
E-value=0.0092743,
Score=91
Ontologies
GO
PANTHER
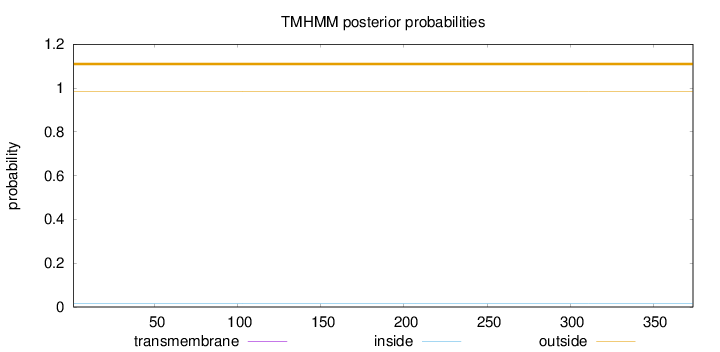
Topology
Subcellular location
Nucleus
Length:
374
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00058
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01483
outside
1 - 374
Population Genetic Test Statistics
Pi
274.65568
Theta
195.363185
Tajima's D
0.497639
CLR
253.318595
CSRT
0.513924303784811
Interpretation
Uncertain
