Gene
KWMTBOMO04154
Pre Gene Modal
BGIBMGA008717
Annotation
PREDICTED:_trafficking_protein_particle_complex_subunit_2_[Papilio_machaon]
Full name
Trafficking protein particle complex subunit 2
+ More
Trafficking protein particle complex subunit 2B
Trafficking protein particle complex subunit 2B
Alternative Name
MBP-1-interacting protein 2A
Sedlin
Sedlin
Location in the cell
Cytoplasmic Reliability : 1.847 Extracellular Reliability : 1.406
Sequence
CDS
ATGGCCGGCACATATTATTTTGTTATAGTCGGACATTCTGACAATCCTATCTTTGAAATGGATTTCATTCCTGTTACTAAAGAAACAAAGAAGGAAGATAACAGACATTTGAACCAGTTCATAGCCCATGCTGCCCTAGATCTAGTGGATGAACACATGTGGAAAGTACCAAATTGTTATTTAAAATCTGTTGACAAATTCAACCAGTGGTTCGTGTCTGCTTTTGTTACAGCAAGCCAGATGAGGTTCATAATAGTTCATGATACAAGAAATGAAGACAGTATAAAAAATTTCTTCACAGATGTCTATGAGTGTTACATTAAATTGATGCTGAATCCATTTTACAAAGAGGATACGCCAATCACAATGCCTGCTTTTGAAAAGAAAGTTCAATTTTTAGGAAAGAAATATTTAACTTAA
Protein
MAGTYYFVIVGHSDNPIFEMDFIPVTKETKKEDNRHLNQFIAHAALDLVDEHMWKVPNCYLKSVDKFNQWFVSAFVTASQMRFIIVHDTRNEDSIKNFFTDVYECYIKLMLNPFYKEDTPITMPAFEKKVQFLGKKYLT
Summary
Description
Prevents ENO1-mediated transcriptional repression and antagonizes ENO1-mediated cell death. May play a role in vesicular transport from endoplasmic reticulum to Golgi (By similarity).
Prevents transcriptional repression and induction of cell death by ENO1. May play a role in vesicular transport from endoplasmic reticulum to Golgi.
Prevents transcriptional repression and induction of cell death by ENO1 (By similarity). May play a role in vesicular transport from endoplasmic reticulum to Golgi.
Prevents transcriptional repression and induction of cell death by ENO1. May play a role in vesicular transport from endoplasmic reticulum to Golgi.
Prevents transcriptional repression and induction of cell death by ENO1 (By similarity). May play a role in vesicular transport from endoplasmic reticulum to Golgi.
Subunit
Part of the multisubunit TRAPP (transport protein particle) complex. Interacts with ENO1, PITX1, SF1, TRAPPC2L and TRAPPC3.
Interacts with PITX1, SF1, and TRAPPC3 (By similarity). Can homodimerize (By similarity). Part of the multisubunit TRAPP (transport protein particle) complex. Interacts with ENO1 and TRAPPC2L.
Can homodimerize. Component of the multisubunit TRAPP (transport protein particle) complex, which includes TRAPPC2, TRAPPC2L, TRAPPC3, TRAPPC3L, TRAPPC4, TRAPPC5, TRAPPC8, TRAPPC9, TRAPPC10, TRAPPC11 and TRAPPC12. Interacts with ENO1, PITX1 and SF1.
Interacts with PITX1, SF1, and TRAPPC3 (By similarity). Can homodimerize (By similarity). Part of the multisubunit TRAPP (transport protein particle) complex. Interacts with ENO1 and TRAPPC2L.
Can homodimerize. Component of the multisubunit TRAPP (transport protein particle) complex, which includes TRAPPC2, TRAPPC2L, TRAPPC3, TRAPPC3L, TRAPPC4, TRAPPC5, TRAPPC8, TRAPPC9, TRAPPC10, TRAPPC11 and TRAPPC12. Interacts with ENO1, PITX1 and SF1.
Miscellaneous
The gene encoding this protein appears to have arisen by retrotransposition of a cDNA from the X-linked locus TRAPPC2. The site of integration of this retrotransposed cDNA appears to lie within an intron of the ZNF547 gene, which provides a promoter and non-coding 5' exon. TRAPPC2 and this protein are indistinguishable at protein level, and both proteins have been identified and functionally characterized.
A paralogous gene encoding an identical protein appears to have arisen by retrotransposition of a cDNA from this locus and to have acquired a promoter and non-coding 5' UTR from the ZNF547 gene.
A paralogous gene encoding an identical protein appears to have arisen by retrotransposition of a cDNA from this locus and to have acquired a promoter and non-coding 5' UTR from the ZNF547 gene.
Similarity
Belongs to the TRAPP small subunits family. Sedlin subfamily.
Keywords
Complete proteome
Cytoplasm
ER-Golgi transport
Nucleus
Reference proteome
Transcription
Transport
Phosphoprotein
Alternative splicing
Disease mutation
Feature
chain Trafficking protein particle complex subunit 2
splice variant In isoform 2 and isoform 3.
sequence variant In SEDT; loss-of-function mutation.
splice variant In isoform 2 and isoform 3.
sequence variant In SEDT; loss-of-function mutation.
Uniprot
H9JGS0
A0A2H1WFV7
I4DKA6
A0A2W1BSZ3
A0A2A4JQA3
A0A212EWR4
+ More
A0A3S2P527 S4P863 A0A0L7L747 A0A067R175 A0A194RD30 A0A0N1PGY9 A0A1B6JBQ9 A0A1B6FDF6 A0A1B6KTV3 A0A146L8N3 A0A069DNT1 E0VPD4 A0A0P4VK01 T1IA30 A0A224Y0F7 A0A0V0G839 A0A1B6DDQ5 A0A1A9ZKG6 A0A1A9UG22 A0A1B0FBU5 H0ZD04 A0A1S2ZD41 U3JPD8 B5KFM3 A0A182LZE4 A0A2R7VRE0 A0A1Y1LQ39 A0A1U7TAR0 A0A1A9WTA7 Q16ZB8 A0A182YHN9 A0A1A9Y051 A0A1B0BC50 A0A0B8RTA1 A0A182RS40 A0A182VWT9 A0A1S3GFZ9 W5PP20 F1SRI0 Q3T0F2 A0A0F7Z927 E3TGK2 A0A1I8PE10 A0A0A0MXP8 A0A2J8RQ25 Q5RES6 A0A287D123 A0A250YA22 A0A286XNW4 A0A384CIG7 A0A341CAP8 A0A2U4AJI9 A0A3Q7N0V3 A0A340XDH8 A0A2Y9FK43 A0A3Q7RAH6 A0A2Y9MWZ2 A0A2U3WN45 A0A3Q7V7T6 A0A2Y9KKL1 E2QV03 G1PL13 A0A0A9Y587 A0A336LU60 A0A084VXW9 A0A2K6CSS0 G3QMD4 A0A2K5XM70 A0A0D9RRH6 A0A2R9BQ93 A0A2K5IBB9 A0A2R8MMX9 A0A2K6UCF0 A0A2J8W4J3 I7G790 F6Z6G0 A0A2J8JLP4 A0A1Y9H2V8 A0A182Q3N6 Q6IBE5 P0DI82 P0DI81 G9KUY4 A0A1I8M8H4 G1RD70 W5MV58 A0A2Y9MRP4 K9IQM0 A0A2Y9ML03 A0A2U3YVX9 U5EL85 A0A1A6HY81 A0A2K6P2H2 A0A240PLN3
A0A3S2P527 S4P863 A0A0L7L747 A0A067R175 A0A194RD30 A0A0N1PGY9 A0A1B6JBQ9 A0A1B6FDF6 A0A1B6KTV3 A0A146L8N3 A0A069DNT1 E0VPD4 A0A0P4VK01 T1IA30 A0A224Y0F7 A0A0V0G839 A0A1B6DDQ5 A0A1A9ZKG6 A0A1A9UG22 A0A1B0FBU5 H0ZD04 A0A1S2ZD41 U3JPD8 B5KFM3 A0A182LZE4 A0A2R7VRE0 A0A1Y1LQ39 A0A1U7TAR0 A0A1A9WTA7 Q16ZB8 A0A182YHN9 A0A1A9Y051 A0A1B0BC50 A0A0B8RTA1 A0A182RS40 A0A182VWT9 A0A1S3GFZ9 W5PP20 F1SRI0 Q3T0F2 A0A0F7Z927 E3TGK2 A0A1I8PE10 A0A0A0MXP8 A0A2J8RQ25 Q5RES6 A0A287D123 A0A250YA22 A0A286XNW4 A0A384CIG7 A0A341CAP8 A0A2U4AJI9 A0A3Q7N0V3 A0A340XDH8 A0A2Y9FK43 A0A3Q7RAH6 A0A2Y9MWZ2 A0A2U3WN45 A0A3Q7V7T6 A0A2Y9KKL1 E2QV03 G1PL13 A0A0A9Y587 A0A336LU60 A0A084VXW9 A0A2K6CSS0 G3QMD4 A0A2K5XM70 A0A0D9RRH6 A0A2R9BQ93 A0A2K5IBB9 A0A2R8MMX9 A0A2K6UCF0 A0A2J8W4J3 I7G790 F6Z6G0 A0A2J8JLP4 A0A1Y9H2V8 A0A182Q3N6 Q6IBE5 P0DI82 P0DI81 G9KUY4 A0A1I8M8H4 G1RD70 W5MV58 A0A2Y9MRP4 K9IQM0 A0A2Y9ML03 A0A2U3YVX9 U5EL85 A0A1A6HY81 A0A2K6P2H2 A0A240PLN3
Pubmed
19121390
22651552
28756777
22118469
23622113
26227816
+ More
24845553 26354079 26823975 26334808 20566863 27129103 20360741 17018643 28004739 17510324 25244985 25476704 20809919 15679890 14681463 20634964 28087693 21993624 24813606 16341006 25401762 24438588 22398555 22722832 17194215 17431167 22002653 25319552 16136131 11181995 11134351 15057824 15489334 10431248 11031107 11805826 19416478 21269460 23186163 14702039 15772651 21525244 25918224 14597397 20498720 11349230 11424925 23236062 25315136 25362486
24845553 26354079 26823975 26334808 20566863 27129103 20360741 17018643 28004739 17510324 25244985 25476704 20809919 15679890 14681463 20634964 28087693 21993624 24813606 16341006 25401762 24438588 22398555 22722832 17194215 17431167 22002653 25319552 16136131 11181995 11134351 15057824 15489334 10431248 11031107 11805826 19416478 21269460 23186163 14702039 15772651 21525244 25918224 14597397 20498720 11349230 11424925 23236062 25315136 25362486
EMBL
BABH01034369
ODYU01008387
SOQ51941.1
AK401724
BAM18346.1
KZ149929
+ More
PZC77461.1 NWSH01000785 PCG74217.1 AGBW02011906 OWR45942.1 RSAL01000020 RVE52585.1 GAIX01006081 JAA86479.1 JTDY01002598 KOB71131.1 KK852818 KDR15689.1 KQ460367 KPJ15522.1 KQ459356 KPJ01266.1 GECU01011121 JAS96585.1 GECZ01032389 GECZ01031363 GECZ01027973 GECZ01025365 GECZ01023297 GECZ01021766 GECZ01013910 GECZ01010307 GECZ01003124 GECZ01002527 GECZ01000935 JAS37380.1 JAS38406.1 JAS41796.1 JAS44404.1 JAS46472.1 JAS48003.1 JAS55859.1 JAS59462.1 JAS66645.1 JAS67242.1 JAS68834.1 GEBQ01025107 JAT14870.1 GDHC01015609 JAQ03020.1 GBGD01003409 JAC85480.1 DS235363 EEB15240.1 GDKW01001242 JAI55353.1 ACPB03012046 GFTR01002343 JAW14083.1 GECL01001849 JAP04275.1 GEDC01013526 JAS23772.1 CCAG010023541 ABQF01021745 AGTO01020707 EF191819 ACH46371.1 AXCM01001996 KK854041 PTY10102.1 GEZM01053531 JAV74025.1 CH477494 EAT39984.1 JXJN01011850 GBSH01002876 JAG66151.1 AMGL01113808 AY610478 AK344004 CU633939 BC102418 GBEX01003661 JAI10899.1 GU589488 ADO29438.1 ABGA01258473 NDHI03003661 PNJ10629.1 PNJ10630.1 CR857440 AGTP01001065 AGTP01001066 GFFW01004365 JAV40423.1 AAKN02052254 AAKN02052255 AAPE02017451 GBHO01016803 GBRD01012766 JAG26801.1 JAG53058.1 UFQS01000194 UFQT01000194 UFQT01000732 SSX01085.1 SSX21465.1 SSX26788.1 ATLV01018209 KE525224 KFB42813.1 CABD030122837 CABD030122838 CABD030122839 AQIB01143495 AJFE02015122 NDHI03003400 PNJ64686.1 PNJ64689.1 PNJ64690.1 AQIA01083425 AB173355 BAE90417.1 JSUE03044944 JU320833 CM001271 AFE64589.1 EHH30458.1 AACZ04014633 NBAG03000445 NBAG03000336 PNI23694.1 PNI23695.1 PNI38635.1 PNI38639.1 AXCN02000007 AK313736 CR456859 CH471135 CH471074 BAG36477.1 CAG33140.1 EAW72495.1 EAW98829.1 EAW98830.1 AF291676 AF058918 AC003002 BC008889 BC032809 AF157065 AF157062 AF157063 AF157064 AC003037 AK310542 DA542477 DB101396 BC016915 BC052618 JP020115 AES08713.1 ADFV01194347 ADFV01194348 AHAT01012851 GABZ01002842 JAA50683.1 GANO01001517 JAB58354.1 LZPO01007952 OBS83206.1
PZC77461.1 NWSH01000785 PCG74217.1 AGBW02011906 OWR45942.1 RSAL01000020 RVE52585.1 GAIX01006081 JAA86479.1 JTDY01002598 KOB71131.1 KK852818 KDR15689.1 KQ460367 KPJ15522.1 KQ459356 KPJ01266.1 GECU01011121 JAS96585.1 GECZ01032389 GECZ01031363 GECZ01027973 GECZ01025365 GECZ01023297 GECZ01021766 GECZ01013910 GECZ01010307 GECZ01003124 GECZ01002527 GECZ01000935 JAS37380.1 JAS38406.1 JAS41796.1 JAS44404.1 JAS46472.1 JAS48003.1 JAS55859.1 JAS59462.1 JAS66645.1 JAS67242.1 JAS68834.1 GEBQ01025107 JAT14870.1 GDHC01015609 JAQ03020.1 GBGD01003409 JAC85480.1 DS235363 EEB15240.1 GDKW01001242 JAI55353.1 ACPB03012046 GFTR01002343 JAW14083.1 GECL01001849 JAP04275.1 GEDC01013526 JAS23772.1 CCAG010023541 ABQF01021745 AGTO01020707 EF191819 ACH46371.1 AXCM01001996 KK854041 PTY10102.1 GEZM01053531 JAV74025.1 CH477494 EAT39984.1 JXJN01011850 GBSH01002876 JAG66151.1 AMGL01113808 AY610478 AK344004 CU633939 BC102418 GBEX01003661 JAI10899.1 GU589488 ADO29438.1 ABGA01258473 NDHI03003661 PNJ10629.1 PNJ10630.1 CR857440 AGTP01001065 AGTP01001066 GFFW01004365 JAV40423.1 AAKN02052254 AAKN02052255 AAPE02017451 GBHO01016803 GBRD01012766 JAG26801.1 JAG53058.1 UFQS01000194 UFQT01000194 UFQT01000732 SSX01085.1 SSX21465.1 SSX26788.1 ATLV01018209 KE525224 KFB42813.1 CABD030122837 CABD030122838 CABD030122839 AQIB01143495 AJFE02015122 NDHI03003400 PNJ64686.1 PNJ64689.1 PNJ64690.1 AQIA01083425 AB173355 BAE90417.1 JSUE03044944 JU320833 CM001271 AFE64589.1 EHH30458.1 AACZ04014633 NBAG03000445 NBAG03000336 PNI23694.1 PNI23695.1 PNI38635.1 PNI38639.1 AXCN02000007 AK313736 CR456859 CH471135 CH471074 BAG36477.1 CAG33140.1 EAW72495.1 EAW98829.1 EAW98830.1 AF291676 AF058918 AC003002 BC008889 BC032809 AF157065 AF157062 AF157063 AF157064 AC003037 AK310542 DA542477 DB101396 BC016915 BC052618 JP020115 AES08713.1 ADFV01194347 ADFV01194348 AHAT01012851 GABZ01002842 JAA50683.1 GANO01001517 JAB58354.1 LZPO01007952 OBS83206.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000037510
UP000027135
+ More
UP000053240 UP000053268 UP000009046 UP000015103 UP000092445 UP000078200 UP000092444 UP000007754 UP000079721 UP000016665 UP000075883 UP000189704 UP000091820 UP000008820 UP000076408 UP000092443 UP000092460 UP000075900 UP000075920 UP000081671 UP000002356 UP000008227 UP000009136 UP000221080 UP000095300 UP000001595 UP000005215 UP000005447 UP000261680 UP000291021 UP000252040 UP000245320 UP000286641 UP000265300 UP000248484 UP000286640 UP000248483 UP000245340 UP000286642 UP000248482 UP000002254 UP000001074 UP000030765 UP000233120 UP000001519 UP000233140 UP000029965 UP000240080 UP000233080 UP000008225 UP000233220 UP000233100 UP000006718 UP000002277 UP000075884 UP000075886 UP000005640 UP000095301 UP000001073 UP000018468 UP000245341 UP000092124 UP000233200 UP000075881
UP000053240 UP000053268 UP000009046 UP000015103 UP000092445 UP000078200 UP000092444 UP000007754 UP000079721 UP000016665 UP000075883 UP000189704 UP000091820 UP000008820 UP000076408 UP000092443 UP000092460 UP000075900 UP000075920 UP000081671 UP000002356 UP000008227 UP000009136 UP000221080 UP000095300 UP000001595 UP000005215 UP000005447 UP000261680 UP000291021 UP000252040 UP000245320 UP000286641 UP000265300 UP000248484 UP000286640 UP000248483 UP000245340 UP000286642 UP000248482 UP000002254 UP000001074 UP000030765 UP000233120 UP000001519 UP000233140 UP000029965 UP000240080 UP000233080 UP000008225 UP000233220 UP000233100 UP000006718 UP000002277 UP000075884 UP000075886 UP000005640 UP000095301 UP000001073 UP000018468 UP000245341 UP000092124 UP000233200 UP000075881
Pfam
PF04628 Sedlin_N
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
H9JGS0
A0A2H1WFV7
I4DKA6
A0A2W1BSZ3
A0A2A4JQA3
A0A212EWR4
+ More
A0A3S2P527 S4P863 A0A0L7L747 A0A067R175 A0A194RD30 A0A0N1PGY9 A0A1B6JBQ9 A0A1B6FDF6 A0A1B6KTV3 A0A146L8N3 A0A069DNT1 E0VPD4 A0A0P4VK01 T1IA30 A0A224Y0F7 A0A0V0G839 A0A1B6DDQ5 A0A1A9ZKG6 A0A1A9UG22 A0A1B0FBU5 H0ZD04 A0A1S2ZD41 U3JPD8 B5KFM3 A0A182LZE4 A0A2R7VRE0 A0A1Y1LQ39 A0A1U7TAR0 A0A1A9WTA7 Q16ZB8 A0A182YHN9 A0A1A9Y051 A0A1B0BC50 A0A0B8RTA1 A0A182RS40 A0A182VWT9 A0A1S3GFZ9 W5PP20 F1SRI0 Q3T0F2 A0A0F7Z927 E3TGK2 A0A1I8PE10 A0A0A0MXP8 A0A2J8RQ25 Q5RES6 A0A287D123 A0A250YA22 A0A286XNW4 A0A384CIG7 A0A341CAP8 A0A2U4AJI9 A0A3Q7N0V3 A0A340XDH8 A0A2Y9FK43 A0A3Q7RAH6 A0A2Y9MWZ2 A0A2U3WN45 A0A3Q7V7T6 A0A2Y9KKL1 E2QV03 G1PL13 A0A0A9Y587 A0A336LU60 A0A084VXW9 A0A2K6CSS0 G3QMD4 A0A2K5XM70 A0A0D9RRH6 A0A2R9BQ93 A0A2K5IBB9 A0A2R8MMX9 A0A2K6UCF0 A0A2J8W4J3 I7G790 F6Z6G0 A0A2J8JLP4 A0A1Y9H2V8 A0A182Q3N6 Q6IBE5 P0DI82 P0DI81 G9KUY4 A0A1I8M8H4 G1RD70 W5MV58 A0A2Y9MRP4 K9IQM0 A0A2Y9ML03 A0A2U3YVX9 U5EL85 A0A1A6HY81 A0A2K6P2H2 A0A240PLN3
A0A3S2P527 S4P863 A0A0L7L747 A0A067R175 A0A194RD30 A0A0N1PGY9 A0A1B6JBQ9 A0A1B6FDF6 A0A1B6KTV3 A0A146L8N3 A0A069DNT1 E0VPD4 A0A0P4VK01 T1IA30 A0A224Y0F7 A0A0V0G839 A0A1B6DDQ5 A0A1A9ZKG6 A0A1A9UG22 A0A1B0FBU5 H0ZD04 A0A1S2ZD41 U3JPD8 B5KFM3 A0A182LZE4 A0A2R7VRE0 A0A1Y1LQ39 A0A1U7TAR0 A0A1A9WTA7 Q16ZB8 A0A182YHN9 A0A1A9Y051 A0A1B0BC50 A0A0B8RTA1 A0A182RS40 A0A182VWT9 A0A1S3GFZ9 W5PP20 F1SRI0 Q3T0F2 A0A0F7Z927 E3TGK2 A0A1I8PE10 A0A0A0MXP8 A0A2J8RQ25 Q5RES6 A0A287D123 A0A250YA22 A0A286XNW4 A0A384CIG7 A0A341CAP8 A0A2U4AJI9 A0A3Q7N0V3 A0A340XDH8 A0A2Y9FK43 A0A3Q7RAH6 A0A2Y9MWZ2 A0A2U3WN45 A0A3Q7V7T6 A0A2Y9KKL1 E2QV03 G1PL13 A0A0A9Y587 A0A336LU60 A0A084VXW9 A0A2K6CSS0 G3QMD4 A0A2K5XM70 A0A0D9RRH6 A0A2R9BQ93 A0A2K5IBB9 A0A2R8MMX9 A0A2K6UCF0 A0A2J8W4J3 I7G790 F6Z6G0 A0A2J8JLP4 A0A1Y9H2V8 A0A182Q3N6 Q6IBE5 P0DI82 P0DI81 G9KUY4 A0A1I8M8H4 G1RD70 W5MV58 A0A2Y9MRP4 K9IQM0 A0A2Y9ML03 A0A2U3YVX9 U5EL85 A0A1A6HY81 A0A2K6P2H2 A0A240PLN3
PDB
2J3W
E-value=1.46182e-51,
Score=506
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Localized in perinuclear granular structures. With evidence from 2 publications.
Perinuclear region Localized in perinuclear granular structures. With evidence from 2 publications.
Nucleus Localized in perinuclear granular structures. With evidence from 2 publications.
Endoplasmic reticulum-Golgi intermediate compartment Localized in perinuclear granular structures. With evidence from 2 publications.
Perinuclear region Localized in perinuclear granular structures. With evidence from 2 publications.
Nucleus Localized in perinuclear granular structures. With evidence from 2 publications.
Endoplasmic reticulum-Golgi intermediate compartment Localized in perinuclear granular structures. With evidence from 2 publications.
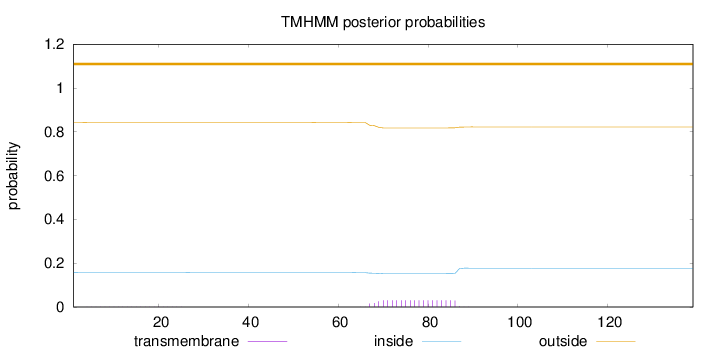
Length:
139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.58572
Exp number, first 60 AAs:
0.00531
Total prob of N-in:
0.15701
outside
1 - 139
Population Genetic Test Statistics
Pi
190.415044
Theta
187.88218
Tajima's D
0.025161
CLR
394.23643
CSRT
0.379581020948953
Interpretation
Uncertain
