Gene
KWMTBOMO04137 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008711
Annotation
PREDICTED:_uncharacterized_protein_LOC105841582_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.762
Sequence
CDS
ATGCGTTGCTTGATAATGTTGTGTGCTTTAATCGCGGTAGCTCTTGCCGCTCCCAACACCATCGGAGATGTCCCGATAGCCCTGCCCTCGCATCCTCGTGGCTGCGTGTGGATCCTCGGAAAATGCTCAAGGAACTGCGAAGAAGGCACCCACAGCTACACGACCGGCTGCCGCCCGAAGATGCCTGAGGCTACATGTGACGTGCCAGTTCCAGTTCAAGAGACACACTCCGTTTGCGACTTCTCCGCGTGCTACTGCGATCCTGGTACAGTCAGGGACACGAAAACGAAGAAATGCGTCAAACTCGAAGACTGCTCGAAGTGA
Protein
MRCLIMLCALIAVALAAPNTIGDVPIALPSHPRGCVWILGKCSRNCEEGTHSYTTGCRPKMPEATCDVPVPVQETHSVCDFSACYCDPGTVRDTKTKKCVKLEDCSK
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
IPR036084
Ser_inhib-like_sf
SUPFAM
SSF57567
SSF57567
ProteinModelPortal
Ontologies
GO
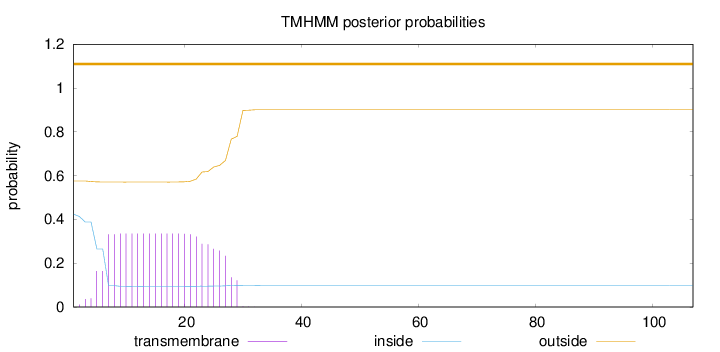
Topology
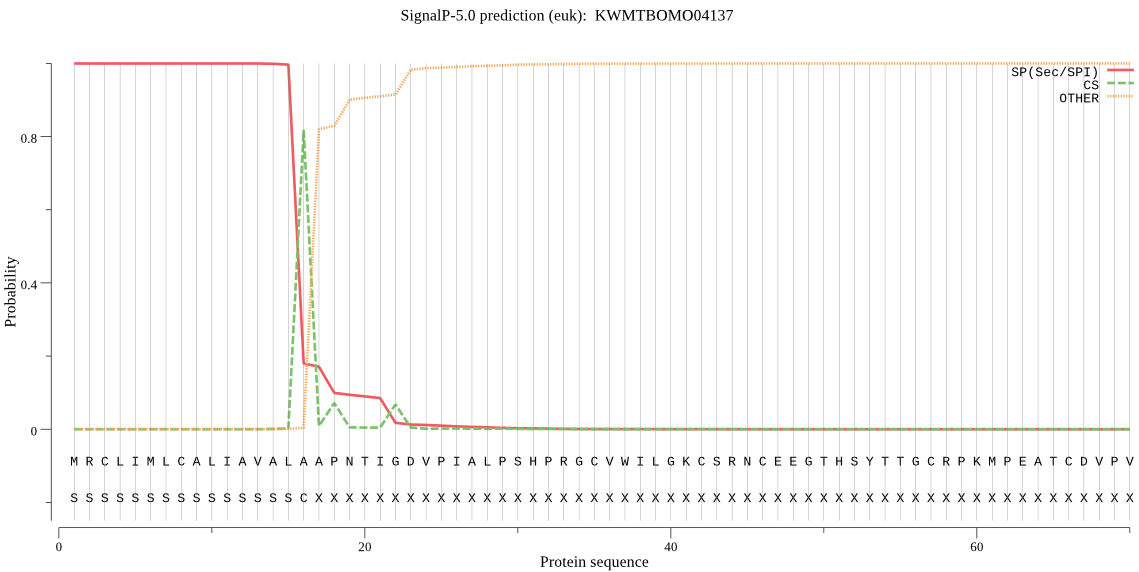
SignalP
Position: 1 - 16,
Likelihood: 0.999615
Length:
107
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.34674
Exp number, first 60 AAs:
7.34674
Total prob of N-in:
0.42428
outside
1 - 107
Population Genetic Test Statistics
Pi
163.965894
Theta
88.116223
Tajima's D
0.3387
CLR
0.819993
CSRT
0.463976801159942
Interpretation
Uncertain
