Gene
KWMTBOMO04132 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008706
Annotation
PREDICTED:_actin-interacting_protein_1_[Amyelois_transitella]
Full name
Actin-interacting protein 1
Alternative Name
Protein flare
Location in the cell
Cytoplasmic Reliability : 1.812 Extracellular Reliability : 1.505
Sequence
CDS
ATGTCTTATTCGAATAAGTATATTTTTGCGGCCCTGCCAAGGACTCAGCGCGGGACGCCGTTAGTTTTAGGCGGTGACCCGAAAGGAAAGAATTTCCTTTACACCAATGGAAATTCTGTTATTATAAGAGACATAGAGAATCCTGCAATTGCGGACGTTTACACGGAGCACTCTTGCCAAGTGAACGTTGCCAAGTACTCTCCTAGCGGATTCTACATTGCATCTGGAGACGCGTCCGGGAAGGTTCGCATCTGGGACACGGTTAACAAGGAACACATCCTAAAGAATGAGTTCCAGCCGATCGGTGGTCCAATAAAGGACATCGCTTGGAGCGCCGACAGCCAACGCATGGTCGTCGTCGGCGAAGGCAGGGAGAGATTCGGTCACGTCTTTATGTCTGAAACCGGTACGTCAGTTGGCGAGATCAGTGGTCAGTCGAAGCCGATAAATTCGGTGGACTTCAGACCCACGAGGCCGTTTCGCATCGTGACCGCCTCAGAGGACAACACCATCGCCGTGTTCGAGGGGCCTCCGTTCAAGTTCAAGTGCACCAAGCAGGAGCATACGCGTTTCGCTCAAGCGGTACGCTACAGTCCAGATGGCAGCCTGTTCGCATCAGCTGGCTTCGATGGCAAGATCTTCCTCTATGACGGAGCCACCTCAGAGCTCAAAGGAGAGATCGGTTCTCCGGCGCACAAGGGCGGCGTTTACGGCATCTCGTGGTCGCCGGACGGTAAACAGTTACTGTCCTGTTCGGGAGACAAGACCTGCGCGATATGGGACATCGAGACGATGCAACGGACCACTCTATTCAACATGGGCAACGCCGTCGAAAACCAACAGGTGTCTTGCCTATGGCAAGGGAAGCATCTCATATCCGTGTCCTTGTCCGGGGTGATCAATTACTTGGATGTCGACAACCCGGACAAGCCGAAGCGAGTGCTGATGGGCCACAACAAGCCGATAACCTGTCTCGCTCTCCACGGCAACGGCAAAACCGTGTACACCGCCAGCCACGACGGCTGCGTCACCGAGTGGAATGTGGACTCGGGCGAAGCTCGTCACGTCGAGGGCCAGGGTCACGGGAACCAGGTCAACGGCATGAGGACGACCAAGGACGGGAGCCTCCTCACCATAGGTATCGACGACACGCTGCGGAAGGCGGAACCCACAGCGGAAGGGGCGACGCCGACGTACACGGGCGCCGCGGTGCCCTTGGGCTCGCAGCCTCGCGCGCTGGACCACGCCGCCGAAGACGACATCACGATCGTCGCCACAGTCAAAGAGCTGGTGGTCTTCGTCGGCGGCAACAAGCAGTCCAGCCTGCCTCTGGCCTACGAGCCGACGTGCGTCGCCATCGACAAGAGCTCGGGGCTGGTCGCCGTCGGCGGCGGCGACAGTCGGGTGCACGTGTACTCGCGCGGGGCGGGCGCGGCGGGCGCGGAGCTGGCGCACCTGGGGCCCGTCACGGACGCGCGCTTCAGTCCGGACTCGCGCTACCTGGTGGCCGGCGACGCTAACCGGAAACTGATACTCTACTCCACCGACGAGTTCAAGCTGGCCCACAACAAGGAGTGGGGCTTCCACACCGCGCGCGTCAACTGCGTGGCCTGGAGCCCGGACTCTAAGCGTGTCGCTTCGGGCTCGCTCGACACCACCATCATCGTCTGGAGCGTTGACCAGCCATCCAAGCACACGATCATTAAGAATGCCCATCCCCAAAGTCAGATAACCGGTCTGACCTGGATCGACGACGAGACCATAGTGTCGACCGGACAGGACGCCAACACGCGGGTCTGGACCGTGCCCAAGGCTTAA
Protein
MSYSNKYIFAALPRTQRGTPLVLGGDPKGKNFLYTNGNSVIIRDIENPAIADVYTEHSCQVNVAKYSPSGFYIASGDASGKVRIWDTVNKEHILKNEFQPIGGPIKDIAWSADSQRMVVVGEGRERFGHVFMSETGTSVGEISGQSKPINSVDFRPTRPFRIVTASEDNTIAVFEGPPFKFKCTKQEHTRFAQAVRYSPDGSLFASAGFDGKIFLYDGATSELKGEIGSPAHKGGVYGISWSPDGKQLLSCSGDKTCAIWDIETMQRTTLFNMGNAVENQQVSCLWQGKHLISVSLSGVINYLDVDNPDKPKRVLMGHNKPITCLALHGNGKTVYTASHDGCVTEWNVDSGEARHVEGQGHGNQVNGMRTTKDGSLLTIGIDDTLRKAEPTAEGATPTYTGAAVPLGSQPRALDHAAEDDITIVATVKELVVFVGGNKQSSLPLAYEPTCVAIDKSSGLVAVGGGDSRVHVYSRGAGAAGAELAHLGPVTDARFSPDSRYLVAGDANRKLILYSTDEFKLAHNKEWGFHTARVNCVAWSPDSKRVASGSLDTTIIVWSVDQPSKHTIIKNAHPQSQITGLTWIDDETIVSTGQDANTRVWTVPKA
Summary
Description
Induces disassembly of actin filaments in conjunction with ADF/cofilin family proteins. Essential for organismal and cell viability. Required for the development of normal wing cell planar polarity.
Similarity
Belongs to the WD repeat AIP1 family.
Keywords
Actin-binding
Alternative splicing
Complete proteome
Cytoplasm
Cytoskeleton
Developmental protein
Reference proteome
Repeat
WD repeat
Feature
chain Actin-interacting protein 1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A1E1VZB5
A0A2A4JVE9
A0A0N1IBD9
H9JGQ9
U5EL22
A0A195B3D8
+ More
A0A195CXM8 A0A158NNL7 A0A026WCT5 F4WQI8 A0A3Q0IY39 A0A195F7X2 A0A067RLI2 A0A151X9G6 A0A0P4VZK9 A0A1B6EZP9 A0A195DLV2 A0A0C9RAF6 A0A0A9W0V8 A0A0A9VRX9 A0A0A9VWC6 A0A0A9VPV3 A0A1B6DMK2 A0A069DVA0 A0A146L7Y1 A0A146LFC8 A0A2M4AL01 A0A0L7QX67 A0A0K8SHM7 A0A0M9AE99 W5J794 A0A088A0M7 A0A182ITH5 A0A3Q0J2I3 A0A023F4T8 A0A232F500 A0A0V0G6W9 A0A1L8E5C1 A0A2A3ERW6 A0A182P974 A0A1L8E5H0 A0A1I9WL82 A0A182FRP4 A0A182Q554 B0X109 A0A182YHM4 A0A182LEY4 A0A182XHW7 A0A182V127 A0A1Q3F2T4 A0A182HSU9 A0A182RGW3 A0A2J7PLM2 Q16P29 A0A023EVG5 A0A2S2PGS4 Q7PRC8 T1HKW3 A0A2H8TDM7 A0A182NRR2 A0A224XDJ1 A0A182MJ51 D6WWM4 J9JRX6 A0A1B0CTT2 W8AUX5 A0A182GA92 N6UD20 A0A084VAQ5 U4UD61 A0A182UAQ5 A0A182U9H2 E2BAM8 A0A182JW63 A0A310SRG5 T1P8S1 B4KVI4 A0A0K8UIE1 A0A034W014 B4LE22 B4IXL6 A0A1I8PCM9 A0A0L0CQH9 K7ITN3 B4MNA9 A0A0A1WNJ1 A0A1J1IJV2 A0A0T6AV18 A0A1W4VIZ8 B3NHE8 B4PHE8 B4HGS4 B4QJ22 Q9VU68 Q29DL4 A0A3B0J829 Q9VU68-2 C8VV50 B3MAM3 A0A1A9Y9A4
A0A195CXM8 A0A158NNL7 A0A026WCT5 F4WQI8 A0A3Q0IY39 A0A195F7X2 A0A067RLI2 A0A151X9G6 A0A0P4VZK9 A0A1B6EZP9 A0A195DLV2 A0A0C9RAF6 A0A0A9W0V8 A0A0A9VRX9 A0A0A9VWC6 A0A0A9VPV3 A0A1B6DMK2 A0A069DVA0 A0A146L7Y1 A0A146LFC8 A0A2M4AL01 A0A0L7QX67 A0A0K8SHM7 A0A0M9AE99 W5J794 A0A088A0M7 A0A182ITH5 A0A3Q0J2I3 A0A023F4T8 A0A232F500 A0A0V0G6W9 A0A1L8E5C1 A0A2A3ERW6 A0A182P974 A0A1L8E5H0 A0A1I9WL82 A0A182FRP4 A0A182Q554 B0X109 A0A182YHM4 A0A182LEY4 A0A182XHW7 A0A182V127 A0A1Q3F2T4 A0A182HSU9 A0A182RGW3 A0A2J7PLM2 Q16P29 A0A023EVG5 A0A2S2PGS4 Q7PRC8 T1HKW3 A0A2H8TDM7 A0A182NRR2 A0A224XDJ1 A0A182MJ51 D6WWM4 J9JRX6 A0A1B0CTT2 W8AUX5 A0A182GA92 N6UD20 A0A084VAQ5 U4UD61 A0A182UAQ5 A0A182U9H2 E2BAM8 A0A182JW63 A0A310SRG5 T1P8S1 B4KVI4 A0A0K8UIE1 A0A034W014 B4LE22 B4IXL6 A0A1I8PCM9 A0A0L0CQH9 K7ITN3 B4MNA9 A0A0A1WNJ1 A0A1J1IJV2 A0A0T6AV18 A0A1W4VIZ8 B3NHE8 B4PHE8 B4HGS4 B4QJ22 Q9VU68 Q29DL4 A0A3B0J829 Q9VU68-2 C8VV50 B3MAM3 A0A1A9Y9A4
Pubmed
26354079
19121390
21347285
24508170
21719571
24845553
+ More
27129103 25401762 26334808 26823975 20920257 23761445 25474469 28648823 27538518 25244985 20966253 17510324 24945155 12364791 14747013 17210077 18362917 19820115 24495485 26483478 23537049 24438588 20798317 25315136 17994087 25348373 26108605 20075255 25830018 17550304 22936249 17565945 10731132 12537572 12537569 15632085
27129103 25401762 26334808 26823975 20920257 23761445 25474469 28648823 27538518 25244985 20966253 17510324 24945155 12364791 14747013 17210077 18362917 19820115 24495485 26483478 23537049 24438588 20798317 25315136 17994087 25348373 26108605 20075255 25830018 17550304 22936249 17565945 10731132 12537572 12537569 15632085
EMBL
GDQN01011030
JAT80024.1
NWSH01000538
PCG75789.1
KQ459356
KPJ01285.1
+ More
BABH01034307 BABH01034308 BABH01034309 GANO01001611 JAB58260.1 KQ976641 KYM78709.1 KQ977141 KYN05415.1 ADTU01021658 KK107311 EZA52859.1 GL888273 EGI63515.1 KQ981744 KYN36287.1 KK852605 KDR20414.1 KQ982383 KYQ56928.1 GDKW01000572 JAI56023.1 GECZ01026243 JAS43526.1 KQ980765 KYN13469.1 GBYB01005255 JAG75022.1 GBHO01045094 JAF98509.1 GBHO01045095 JAF98508.1 GBHO01045096 JAF98507.1 GBHO01045097 JAF98506.1 GEDC01010448 JAS26850.1 GBGD01000891 JAC87998.1 GDHC01014924 JAQ03705.1 GDHC01012903 JAQ05726.1 GGFK01008140 MBW41461.1 KQ414705 KOC63212.1 GBRD01013222 JAG52604.1 KQ435687 KOX81379.1 ADMH02001952 ETN60327.1 GBBI01002236 JAC16476.1 NNAY01000998 OXU25558.1 GECL01002437 JAP03687.1 GFDF01000137 JAV13947.1 KZ288191 PBC34505.1 GFDF01000136 JAV13948.1 KU932266 APA33902.1 AXCN02000008 DS232250 EDS38401.1 GFDL01013182 JAV21863.1 APCN01000365 NEVH01024424 PNF17231.1 CH477797 EAT36116.1 GAPW01000601 JAC12997.1 GGMR01015779 MBY28398.1 AAAB01008859 EAA07462.6 ACPB03020751 ACPB03020752 ACPB03020753 ACPB03020754 ACPB03020755 ACPB03020756 ACPB03020757 GFXV01000365 MBW12170.1 GFTR01007372 JAW09054.1 AXCM01004352 KQ971361 EFA08118.1 ABLF02021242 AJWK01027950 AJWK01027951 GAMC01016519 GAMC01016518 JAB90037.1 JXUM01008452 JXUM01008453 JXUM01008454 KQ560261 KXJ83493.1 APGK01040103 KB740975 ENN76547.1 ATLV01004135 KE524190 KFB35049.1 KB632267 ERL90992.1 GL446793 EFN87254.1 KQ759893 OAD62099.1 KA645079 AFP59708.1 CH933809 EDW19455.1 GDHF01025865 JAI26449.1 GAKP01011819 GAKP01011818 JAC47133.1 CH940647 EDW70065.1 CH916366 EDV97478.1 JRES01000045 KNC34623.1 CH963847 EDW72618.1 GBXI01014309 JAC99982.1 CVRI01000050 CRK99342.1 LJIG01022745 KRT78936.1 CH954178 EDV51674.1 CM000159 EDW94409.1 CH480815 EDW41382.1 CM000363 CM002912 EDX10353.1 KMY99386.1 AE014296 AY061340 CH379070 EAL30400.1 OUUW01000002 SPP76373.1 BT099796 ACV91633.1 CH902618 EDV41310.1
BABH01034307 BABH01034308 BABH01034309 GANO01001611 JAB58260.1 KQ976641 KYM78709.1 KQ977141 KYN05415.1 ADTU01021658 KK107311 EZA52859.1 GL888273 EGI63515.1 KQ981744 KYN36287.1 KK852605 KDR20414.1 KQ982383 KYQ56928.1 GDKW01000572 JAI56023.1 GECZ01026243 JAS43526.1 KQ980765 KYN13469.1 GBYB01005255 JAG75022.1 GBHO01045094 JAF98509.1 GBHO01045095 JAF98508.1 GBHO01045096 JAF98507.1 GBHO01045097 JAF98506.1 GEDC01010448 JAS26850.1 GBGD01000891 JAC87998.1 GDHC01014924 JAQ03705.1 GDHC01012903 JAQ05726.1 GGFK01008140 MBW41461.1 KQ414705 KOC63212.1 GBRD01013222 JAG52604.1 KQ435687 KOX81379.1 ADMH02001952 ETN60327.1 GBBI01002236 JAC16476.1 NNAY01000998 OXU25558.1 GECL01002437 JAP03687.1 GFDF01000137 JAV13947.1 KZ288191 PBC34505.1 GFDF01000136 JAV13948.1 KU932266 APA33902.1 AXCN02000008 DS232250 EDS38401.1 GFDL01013182 JAV21863.1 APCN01000365 NEVH01024424 PNF17231.1 CH477797 EAT36116.1 GAPW01000601 JAC12997.1 GGMR01015779 MBY28398.1 AAAB01008859 EAA07462.6 ACPB03020751 ACPB03020752 ACPB03020753 ACPB03020754 ACPB03020755 ACPB03020756 ACPB03020757 GFXV01000365 MBW12170.1 GFTR01007372 JAW09054.1 AXCM01004352 KQ971361 EFA08118.1 ABLF02021242 AJWK01027950 AJWK01027951 GAMC01016519 GAMC01016518 JAB90037.1 JXUM01008452 JXUM01008453 JXUM01008454 KQ560261 KXJ83493.1 APGK01040103 KB740975 ENN76547.1 ATLV01004135 KE524190 KFB35049.1 KB632267 ERL90992.1 GL446793 EFN87254.1 KQ759893 OAD62099.1 KA645079 AFP59708.1 CH933809 EDW19455.1 GDHF01025865 JAI26449.1 GAKP01011819 GAKP01011818 JAC47133.1 CH940647 EDW70065.1 CH916366 EDV97478.1 JRES01000045 KNC34623.1 CH963847 EDW72618.1 GBXI01014309 JAC99982.1 CVRI01000050 CRK99342.1 LJIG01022745 KRT78936.1 CH954178 EDV51674.1 CM000159 EDW94409.1 CH480815 EDW41382.1 CM000363 CM002912 EDX10353.1 KMY99386.1 AE014296 AY061340 CH379070 EAL30400.1 OUUW01000002 SPP76373.1 BT099796 ACV91633.1 CH902618 EDV41310.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000078540
UP000078542
UP000005205
+ More
UP000053097 UP000007755 UP000079169 UP000078541 UP000027135 UP000075809 UP000078492 UP000053825 UP000053105 UP000000673 UP000005203 UP000075880 UP000215335 UP000242457 UP000075885 UP000069272 UP000075886 UP000002320 UP000076408 UP000075882 UP000076407 UP000075903 UP000075840 UP000075900 UP000235965 UP000008820 UP000007062 UP000015103 UP000075884 UP000075883 UP000007266 UP000007819 UP000092461 UP000069940 UP000249989 UP000019118 UP000030765 UP000030742 UP000075902 UP000008237 UP000075881 UP000095301 UP000009192 UP000008792 UP000001070 UP000095300 UP000037069 UP000002358 UP000007798 UP000183832 UP000192221 UP000008711 UP000002282 UP000001292 UP000000304 UP000000803 UP000001819 UP000268350 UP000007801 UP000092443
UP000053097 UP000007755 UP000079169 UP000078541 UP000027135 UP000075809 UP000078492 UP000053825 UP000053105 UP000000673 UP000005203 UP000075880 UP000215335 UP000242457 UP000075885 UP000069272 UP000075886 UP000002320 UP000076408 UP000075882 UP000076407 UP000075903 UP000075840 UP000075900 UP000235965 UP000008820 UP000007062 UP000015103 UP000075884 UP000075883 UP000007266 UP000007819 UP000092461 UP000069940 UP000249989 UP000019118 UP000030765 UP000030742 UP000075902 UP000008237 UP000075881 UP000095301 UP000009192 UP000008792 UP000001070 UP000095300 UP000037069 UP000002358 UP000007798 UP000183832 UP000192221 UP000008711 UP000002282 UP000001292 UP000000304 UP000000803 UP000001819 UP000268350 UP000007801 UP000092443
Pfam
Interpro
IPR020472
G-protein_beta_WD-40_rep
+ More
IPR019775 WD40_repeat_CS
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR013979 TIF_beta_prop-like
IPR031544 WD40-like
IPR024977 Apc4_WD40_dom
IPR002350 Kazal_dom
IPR036058 Kazal_dom_sf
IPR012346 p53/RUNT-type_TF_DNA-bd_sf
IPR006966 Peroxin-3
IPR000040 AML1_Runt
IPR013524 Runt_dom
IPR008967 p53-like_TF_DNA-bd
IPR011043 Gal_Oxase/kelch_b-propeller
IPR019775 WD40_repeat_CS
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR013979 TIF_beta_prop-like
IPR031544 WD40-like
IPR024977 Apc4_WD40_dom
IPR002350 Kazal_dom
IPR036058 Kazal_dom_sf
IPR012346 p53/RUNT-type_TF_DNA-bd_sf
IPR006966 Peroxin-3
IPR000040 AML1_Runt
IPR013524 Runt_dom
IPR008967 p53-like_TF_DNA-bd
IPR011043 Gal_Oxase/kelch_b-propeller
SUPFAM
Gene 3D
ProteinModelPortal
A0A1E1VZB5
A0A2A4JVE9
A0A0N1IBD9
H9JGQ9
U5EL22
A0A195B3D8
+ More
A0A195CXM8 A0A158NNL7 A0A026WCT5 F4WQI8 A0A3Q0IY39 A0A195F7X2 A0A067RLI2 A0A151X9G6 A0A0P4VZK9 A0A1B6EZP9 A0A195DLV2 A0A0C9RAF6 A0A0A9W0V8 A0A0A9VRX9 A0A0A9VWC6 A0A0A9VPV3 A0A1B6DMK2 A0A069DVA0 A0A146L7Y1 A0A146LFC8 A0A2M4AL01 A0A0L7QX67 A0A0K8SHM7 A0A0M9AE99 W5J794 A0A088A0M7 A0A182ITH5 A0A3Q0J2I3 A0A023F4T8 A0A232F500 A0A0V0G6W9 A0A1L8E5C1 A0A2A3ERW6 A0A182P974 A0A1L8E5H0 A0A1I9WL82 A0A182FRP4 A0A182Q554 B0X109 A0A182YHM4 A0A182LEY4 A0A182XHW7 A0A182V127 A0A1Q3F2T4 A0A182HSU9 A0A182RGW3 A0A2J7PLM2 Q16P29 A0A023EVG5 A0A2S2PGS4 Q7PRC8 T1HKW3 A0A2H8TDM7 A0A182NRR2 A0A224XDJ1 A0A182MJ51 D6WWM4 J9JRX6 A0A1B0CTT2 W8AUX5 A0A182GA92 N6UD20 A0A084VAQ5 U4UD61 A0A182UAQ5 A0A182U9H2 E2BAM8 A0A182JW63 A0A310SRG5 T1P8S1 B4KVI4 A0A0K8UIE1 A0A034W014 B4LE22 B4IXL6 A0A1I8PCM9 A0A0L0CQH9 K7ITN3 B4MNA9 A0A0A1WNJ1 A0A1J1IJV2 A0A0T6AV18 A0A1W4VIZ8 B3NHE8 B4PHE8 B4HGS4 B4QJ22 Q9VU68 Q29DL4 A0A3B0J829 Q9VU68-2 C8VV50 B3MAM3 A0A1A9Y9A4
A0A195CXM8 A0A158NNL7 A0A026WCT5 F4WQI8 A0A3Q0IY39 A0A195F7X2 A0A067RLI2 A0A151X9G6 A0A0P4VZK9 A0A1B6EZP9 A0A195DLV2 A0A0C9RAF6 A0A0A9W0V8 A0A0A9VRX9 A0A0A9VWC6 A0A0A9VPV3 A0A1B6DMK2 A0A069DVA0 A0A146L7Y1 A0A146LFC8 A0A2M4AL01 A0A0L7QX67 A0A0K8SHM7 A0A0M9AE99 W5J794 A0A088A0M7 A0A182ITH5 A0A3Q0J2I3 A0A023F4T8 A0A232F500 A0A0V0G6W9 A0A1L8E5C1 A0A2A3ERW6 A0A182P974 A0A1L8E5H0 A0A1I9WL82 A0A182FRP4 A0A182Q554 B0X109 A0A182YHM4 A0A182LEY4 A0A182XHW7 A0A182V127 A0A1Q3F2T4 A0A182HSU9 A0A182RGW3 A0A2J7PLM2 Q16P29 A0A023EVG5 A0A2S2PGS4 Q7PRC8 T1HKW3 A0A2H8TDM7 A0A182NRR2 A0A224XDJ1 A0A182MJ51 D6WWM4 J9JRX6 A0A1B0CTT2 W8AUX5 A0A182GA92 N6UD20 A0A084VAQ5 U4UD61 A0A182UAQ5 A0A182U9H2 E2BAM8 A0A182JW63 A0A310SRG5 T1P8S1 B4KVI4 A0A0K8UIE1 A0A034W014 B4LE22 B4IXL6 A0A1I8PCM9 A0A0L0CQH9 K7ITN3 B4MNA9 A0A0A1WNJ1 A0A1J1IJV2 A0A0T6AV18 A0A1W4VIZ8 B3NHE8 B4PHE8 B4HGS4 B4QJ22 Q9VU68 Q29DL4 A0A3B0J829 Q9VU68-2 C8VV50 B3MAM3 A0A1A9Y9A4
Ontologies
GO
GO:0003677
GO:0005634
GO:0007031
GO:0005779
GO:0003700
GO:0005524
GO:0030864
GO:0042643
GO:0051015
GO:0030042
GO:0030836
GO:0040011
GO:0045214
GO:0035317
GO:0005856
GO:0030834
GO:0007298
GO:0034316
GO:0001736
GO:0003743
GO:0003779
GO:0005737
GO:0030833
GO:0005515
GO:0003723
GO:0009451
GO:0009982
GO:0006508
GO:0042555
GO:0006260
GO:0016021
GO:0004672
GO:0048384
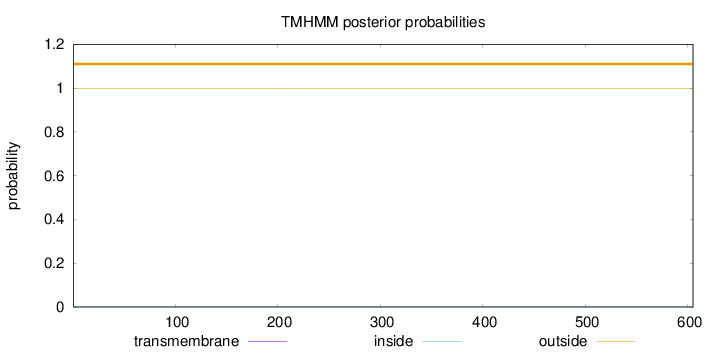
Topology
Subcellular location
Length:
605
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000620000000000001
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00108
outside
1 - 605
Population Genetic Test Statistics
Pi
249.198942
Theta
169.341026
Tajima's D
0.425004
CLR
0.698883
CSRT
0.487875606219689
Interpretation
Uncertain
