Gene
KWMTBOMO04124
Pre Gene Modal
BGIBMGA005185
Annotation
PREDICTED:_uncharacterized_protein_LOC101746336_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.366
Sequence
CDS
ATGTCGCAAAAACAAGTTAGAAAACGTGGAGCAAATTACACTCAAGAGGAAAAAACATTGCTGCTGGATCTCATCATGCAATATAAAGATATTGTGGAAAGTAAACGAACAGGTGGCATTATTATCCAAAAGAAAAAGGAGGTTTGGAGAACTATAACCTCAAAATATAATAGCAGCTGCAGTAGTGGTCCCAGGGATGTTGACCAGCTGAAAGCTTTATATGACAATATGAAGCAAAAATCTCGCAAAGAAATTGGTGAAAAAAATGACTCCGGTATTCCGTTCGAATTTATTAACCTGGCGGCCGATCCCGTCACTTTATTTAAAATACCATTTATAAGAGCTAAATTGGACAACTCATATCCTGGTTCGGTTCTAAAATGA
Protein
MSQKQVRKRGANYTQEEKTLLLDLIMQYKDIVESKRTGGIIIQKKKEVWRTITSKYNSSCSSGPRDVDQLKALYDNMKQKSRKEIGEKNDSGIPFEFINLAADPVTLFKIPFIRAKLDNSYPGSVLK
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
Pubmed
EMBL
ODYU01005534
SOQ46478.1
KZ150042
PZC74542.1
KQ971356
EFA09122.1
+ More
ABLF02013368 GEZM01000299 GEZM01000298 JAV98436.1 ABLF02003707 KQ980775 KYN13234.1 LJIG01000044 KRT86896.1 ODYU01009057 SOQ53198.1 PYGN01000342 PSN48118.1 ABLF02029655 OUUW01000011 SPP86277.1 SPP86276.1 CH379063 KRT06094.1 KRT06097.1 ABLF02011428 KRT06093.1 KRT06096.1 ABLF02018378 KRT06095.1 CH479190 EDW25891.1 GDHC01000687 JAQ17942.1 EDY72244.2 GBHO01011257 GBHO01011254 JAG32347.1 JAG32350.1
ABLF02013368 GEZM01000299 GEZM01000298 JAV98436.1 ABLF02003707 KQ980775 KYN13234.1 LJIG01000044 KRT86896.1 ODYU01009057 SOQ53198.1 PYGN01000342 PSN48118.1 ABLF02029655 OUUW01000011 SPP86277.1 SPP86276.1 CH379063 KRT06094.1 KRT06097.1 ABLF02011428 KRT06093.1 KRT06096.1 ABLF02018378 KRT06095.1 CH479190 EDW25891.1 GDHC01000687 JAQ17942.1 EDY72244.2 GBHO01011257 GBHO01011254 JAG32347.1 JAG32350.1
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
Ontologies
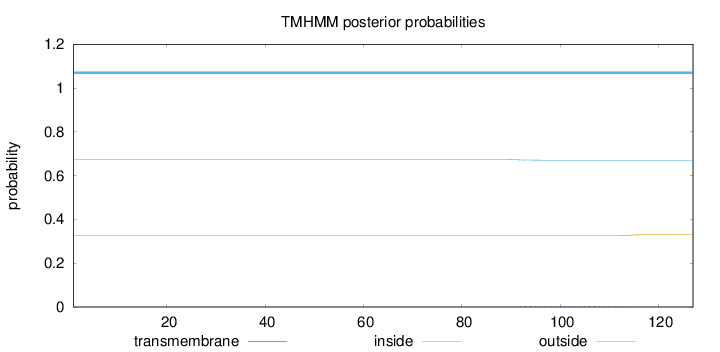
Topology
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09704
Exp number, first 60 AAs:
0
Total prob of N-in:
0.67450
inside
1 - 127
Population Genetic Test Statistics
Pi
226.46269
Theta
170.679546
Tajima's D
0.852129
CLR
0.28586
CSRT
0.618319084045798
Interpretation
Uncertain
