Pre Gene Modal
BGIBMGA008699
Annotation
PREDICTED:_UPF0160_protein_MYG1?_mitochondrial_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.058
Sequence
CDS
ATGAAGATTGGAACTCACGACGGAGTATTCCACTGTGATGAAGTCCTAGCTTGCTTTATGCTGAAGAATCTTCCTCAATACAAAGATGCTGAGATCATCCGCACAAGAGATCTAAATAAGCTGAATGACTGTGATATAGTTGTTGATGTAGGCAGTGTTTTTGACCATGAAAAGAAGCGTTATGATCATCACCAAGCCGGTTTTAATGAGACCTTGAGTACCTTGAGGCCTGAACTCGGAGGCAGCTATAAAATTAAACTTAGTTCAGCAGGCCTTGTGTATGCTTACTATGGAGAGGACATCATCCAGCAACTCAAAGAAGAGTCTACCTCCTTGACAAATGAAGACTTGAAGTTAATTTACAAGAAAGTTTATGAATCTTTCATTCAAGAGATTGATGCTATTGATAACGGAATTCCAATGACAGAGGAACAACCGAAATATGACATTCATACTCACTTAAGTAACAGAGTGAAAAGACTGAACCCAGAATGGAATTCAACTCAGGAAACCAATGTAGATGAATTCTTCAATAAAGCAATGGCTTTAGTTAGTGAAGATTTCCTATATTTTACAAGATACTACATAAACACTTGGTTTCCAGCCCGAAATTTGGTCAGGAAAGCTATTGATAATCGTTTTCAAATACACAAGTCAGGAGAGATTGTGGTATTCAATGAAAGGTTTCCTTGGAAATATCATTTGTTTGATTTGGAACAAGAATTGGGGATTGAGCCGCAAATCAAATATGTGCTATTCAATGATAAACCAAAATCTTGGAGAGTTCAAGCTGTTCCAATTGATCCTATGAGTTTTGTTCTTAGGAAAGCATTACATAGTGCATGGCGTGGAATCCGTGACGATAATCTCAGTTCAGTCTCTGGGATTGAAGGATGTATTTTCTGCCACAGCAATGGTTTCATTGGTGGTAATACCACATTGGAGGGAGCTTTGAAAATGGCCATTGCCAGCCTAGAAGCCAACGAATGA
Protein
MKIGTHDGVFHCDEVLACFMLKNLPQYKDAEIIRTRDLNKLNDCDIVVDVGSVFDHEKKRYDHHQAGFNETLSTLRPELGGSYKIKLSSAGLVYAYYGEDIIQQLKEESTSLTNEDLKLIYKKVYESFIQEIDAIDNGIPMTEEQPKYDIHTHLSNRVKRLNPEWNSTQETNVDEFFNKAMALVSEDFLYFTRYYINTWFPARNLVRKAIDNRFQIHKSGEIVVFNERFPWKYHLFDLEQELGIEPQIKYVLFNDKPKSWRVQAVPIDPMSFVLRKALHSAWRGIRDDNLSSVSGIEGCIFCHSNGFIGGNTTLEGALKMAIASLEANE
Summary
Uniprot
A0A2A4J1N7
A0A1E1WCX0
A0A2H1VTJ1
A0A0N0P9K5
A0A3S2TFW6
A0A212F6A6
+ More
A0A2A4J316 A0A194RKW5 D6WP32 A0A1E1WQN8 A0A182P710 A0A0T6BA06 A0A182U8B0 Q7QC19 U5EVU0 A0A182LFA3 A0A1Y9J005 A0A182VX73 A0A182HT64 A0A182UQL5 A0A182K7C7 N6TNI3 A0A1W4X8Y9 A0A182T8M8 A0A1W4XJF3 A0A1W4XJ63 T1EA83 A0A182YKG3 Q172K4 A0A182J6G6 B0XKE1 A0A182MEA5 A0A182QRS7 A0A182NA65 A0A336LPI0 A0A2M4APH7 E0VKY8 W5JK47 Q172K5 A0A1J1IHR0 A0A182H0R0 A0A1Q3F1L4 A0A182H139 A0A023ERL7 A0A084VSD6 A0A1B0D999 A0A1Y1MX31 A0A1Y1MU12 A0A2T7PJ00 A0A0K8TR04 A0A067RCM3 A0A1L8E032 R4G836 A0A0P7ZDL0 K7J738 A0A2S2Q1Y5 W5ME83 A0A2J7PQK2 A0A1W4ZAJ3 A0A2J7PQG3 A0A1B6DLN8 A0A3B3R8M2 A0A195D5R8 A9UV55 A0A0A1WJ22 V4A4D9 R7V123 A0A0A1WWD0 Q4RX01 H3DBH6 A0A170XUD1 A0A3Q3MIC2 A0A195AZH1 C3XWS9 A0A1W2WJA3 A0A1W2WR78 H2ZNV1 E9I8N9 A0A195FWN8 F4WD91 A7S559 F6ZCS4 C3XWP2 A0A2H8TFE1 A0A2U9BF80 W8C9P3 A0A195E241 A0A3Q3LYS5 A0A2S2N8G4 A0A158NHN3 A0A0V0G881 A0A3Q3ECQ1 I3JC73 A0A3Q4B731 H2LKA5 A0A1B0BSA6 A0A154PA46 H3B2C3 A0A224XSQ5
A0A2A4J316 A0A194RKW5 D6WP32 A0A1E1WQN8 A0A182P710 A0A0T6BA06 A0A182U8B0 Q7QC19 U5EVU0 A0A182LFA3 A0A1Y9J005 A0A182VX73 A0A182HT64 A0A182UQL5 A0A182K7C7 N6TNI3 A0A1W4X8Y9 A0A182T8M8 A0A1W4XJF3 A0A1W4XJ63 T1EA83 A0A182YKG3 Q172K4 A0A182J6G6 B0XKE1 A0A182MEA5 A0A182QRS7 A0A182NA65 A0A336LPI0 A0A2M4APH7 E0VKY8 W5JK47 Q172K5 A0A1J1IHR0 A0A182H0R0 A0A1Q3F1L4 A0A182H139 A0A023ERL7 A0A084VSD6 A0A1B0D999 A0A1Y1MX31 A0A1Y1MU12 A0A2T7PJ00 A0A0K8TR04 A0A067RCM3 A0A1L8E032 R4G836 A0A0P7ZDL0 K7J738 A0A2S2Q1Y5 W5ME83 A0A2J7PQK2 A0A1W4ZAJ3 A0A2J7PQG3 A0A1B6DLN8 A0A3B3R8M2 A0A195D5R8 A9UV55 A0A0A1WJ22 V4A4D9 R7V123 A0A0A1WWD0 Q4RX01 H3DBH6 A0A170XUD1 A0A3Q3MIC2 A0A195AZH1 C3XWS9 A0A1W2WJA3 A0A1W2WR78 H2ZNV1 E9I8N9 A0A195FWN8 F4WD91 A7S559 F6ZCS4 C3XWP2 A0A2H8TFE1 A0A2U9BF80 W8C9P3 A0A195E241 A0A3Q3LYS5 A0A2S2N8G4 A0A158NHN3 A0A0V0G881 A0A3Q3ECQ1 I3JC73 A0A3Q4B731 H2LKA5 A0A1B0BSA6 A0A154PA46 H3B2C3 A0A224XSQ5
Pubmed
26354079
22118469
18362917
19820115
12364791
14747013
+ More
17210077 20966253 23537049 25244985 17510324 20566863 20920257 23761445 26483478 24945155 24438588 28004739 26369729 24845553 20075255 29240929 18273011 25830018 23254933 15496914 18563158 21282665 21719571 17615350 12481130 15114417 24495485 21347285 25186727 17554307 9215903
17210077 20966253 23537049 25244985 17510324 20566863 20920257 23761445 26483478 24945155 24438588 28004739 26369729 24845553 20075255 29240929 18273011 25830018 23254933 15496914 18563158 21282665 21719571 17615350 12481130 15114417 24495485 21347285 25186727 17554307 9215903
EMBL
NWSH01003588
PCG66077.1
GDQN01006231
JAT84823.1
ODYU01004340
SOQ44098.1
+ More
KQ459356 KPJ01289.1 RSAL01000183 RVE44872.1 AGBW02010070 OWR49263.1 PCG66078.1 KQ460075 KPJ17970.1 KQ971352 EFA07253.1 GDQN01001734 JAT89320.1 LJIG01002778 KRT84162.1 AAAB01008859 EAA08054.3 GANO01003259 JAB56612.1 APCN01000420 APGK01057815 KB741282 KB631684 ENN70810.1 ERL85369.1 GAMD01000662 JAB00929.1 CH477435 EAT40960.1 DS233805 EDS31662.1 AXCM01000752 AXCN02000216 UFQT01000038 SSX18469.1 GGFK01009374 MBW42695.1 DS235255 EEB14044.1 ADMH02001206 ETN63683.1 EAT40959.1 CVRI01000053 CRK99789.1 JXUM01101736 KQ564671 KXJ71925.1 GFDL01013595 JAV21450.1 JXUM01102433 KQ564727 KXJ71818.1 GAPW01001753 JAC11845.1 ATLV01015922 KE525044 KFB40880.1 AJVK01004286 GEZM01021142 JAV89120.1 GEZM01021141 JAV89121.1 PZQS01000003 PVD33405.1 GDAI01001030 JAI16573.1 KK852726 KDR17656.1 GFDF01002043 JAV12041.1 ACPB03007616 GAHY01001657 JAA75853.1 JARO02000283 KPP79134.1 AAZX01002971 GGMS01002594 MBY71797.1 AHAT01018765 AHAT01018766 NEVH01022638 PNF18576.1 PNF18578.1 GEDC01010721 JAS26577.1 KQ976870 KYN07779.1 CH991546 EDQ90835.1 GBXI01015767 JAC98524.1 KB202237 ESO91567.1 AMQN01000816 KB296161 ELU12172.1 GBXI01010943 JAD03349.1 CAAE01014979 CAG07081.1 GEMB01004028 JAR99233.1 KQ976694 KYM77601.1 GG666471 EEN67189.1 GL761591 EFZ23073.1 KQ981208 KYN44853.1 GL888086 EGI67818.1 DS469581 EDO41186.1 EAAA01002750 EAAA01002751 EEN67152.1 GFXV01000999 MBW12804.1 CP026248 AWP02577.1 GAMC01006461 GAMC01006459 JAC00095.1 KQ979763 KYN19208.1 GGMR01000904 MBY13523.1 ADTU01015734 GECL01002647 JAP03477.1 AERX01027111 AERX01027112 AERX01027113 AERX01027114 JXJN01019614 KQ434857 KZC08779.1 AFYH01001162 AFYH01001163 AFYH01001164 AFYH01001165 AFYH01001166 AFYH01001167 AFYH01001168 AFYH01001169 GFTR01005273 JAW11153.1
KQ459356 KPJ01289.1 RSAL01000183 RVE44872.1 AGBW02010070 OWR49263.1 PCG66078.1 KQ460075 KPJ17970.1 KQ971352 EFA07253.1 GDQN01001734 JAT89320.1 LJIG01002778 KRT84162.1 AAAB01008859 EAA08054.3 GANO01003259 JAB56612.1 APCN01000420 APGK01057815 KB741282 KB631684 ENN70810.1 ERL85369.1 GAMD01000662 JAB00929.1 CH477435 EAT40960.1 DS233805 EDS31662.1 AXCM01000752 AXCN02000216 UFQT01000038 SSX18469.1 GGFK01009374 MBW42695.1 DS235255 EEB14044.1 ADMH02001206 ETN63683.1 EAT40959.1 CVRI01000053 CRK99789.1 JXUM01101736 KQ564671 KXJ71925.1 GFDL01013595 JAV21450.1 JXUM01102433 KQ564727 KXJ71818.1 GAPW01001753 JAC11845.1 ATLV01015922 KE525044 KFB40880.1 AJVK01004286 GEZM01021142 JAV89120.1 GEZM01021141 JAV89121.1 PZQS01000003 PVD33405.1 GDAI01001030 JAI16573.1 KK852726 KDR17656.1 GFDF01002043 JAV12041.1 ACPB03007616 GAHY01001657 JAA75853.1 JARO02000283 KPP79134.1 AAZX01002971 GGMS01002594 MBY71797.1 AHAT01018765 AHAT01018766 NEVH01022638 PNF18576.1 PNF18578.1 GEDC01010721 JAS26577.1 KQ976870 KYN07779.1 CH991546 EDQ90835.1 GBXI01015767 JAC98524.1 KB202237 ESO91567.1 AMQN01000816 KB296161 ELU12172.1 GBXI01010943 JAD03349.1 CAAE01014979 CAG07081.1 GEMB01004028 JAR99233.1 KQ976694 KYM77601.1 GG666471 EEN67189.1 GL761591 EFZ23073.1 KQ981208 KYN44853.1 GL888086 EGI67818.1 DS469581 EDO41186.1 EAAA01002750 EAAA01002751 EEN67152.1 GFXV01000999 MBW12804.1 CP026248 AWP02577.1 GAMC01006461 GAMC01006459 JAC00095.1 KQ979763 KYN19208.1 GGMR01000904 MBY13523.1 ADTU01015734 GECL01002647 JAP03477.1 AERX01027111 AERX01027112 AERX01027113 AERX01027114 JXJN01019614 KQ434857 KZC08779.1 AFYH01001162 AFYH01001163 AFYH01001164 AFYH01001165 AFYH01001166 AFYH01001167 AFYH01001168 AFYH01001169 GFTR01005273 JAW11153.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
UP000007266
+ More
UP000075885 UP000075902 UP000007062 UP000075882 UP000076407 UP000075920 UP000075840 UP000075903 UP000075881 UP000019118 UP000030742 UP000192223 UP000075901 UP000076408 UP000008820 UP000075880 UP000002320 UP000075883 UP000075886 UP000075884 UP000009046 UP000000673 UP000183832 UP000069940 UP000249989 UP000030765 UP000092462 UP000245119 UP000027135 UP000015103 UP000034805 UP000002358 UP000018468 UP000235965 UP000192224 UP000261540 UP000078542 UP000001357 UP000030746 UP000014760 UP000007303 UP000261660 UP000078540 UP000001554 UP000007875 UP000078541 UP000007755 UP000001593 UP000008144 UP000246464 UP000078492 UP000261640 UP000005205 UP000264820 UP000005207 UP000261620 UP000001038 UP000092460 UP000076502 UP000008672
UP000075885 UP000075902 UP000007062 UP000075882 UP000076407 UP000075920 UP000075840 UP000075903 UP000075881 UP000019118 UP000030742 UP000192223 UP000075901 UP000076408 UP000008820 UP000075880 UP000002320 UP000075883 UP000075886 UP000075884 UP000009046 UP000000673 UP000183832 UP000069940 UP000249989 UP000030765 UP000092462 UP000245119 UP000027135 UP000015103 UP000034805 UP000002358 UP000018468 UP000235965 UP000192224 UP000261540 UP000078542 UP000001357 UP000030746 UP000014760 UP000007303 UP000261660 UP000078540 UP000001554 UP000007875 UP000078541 UP000007755 UP000001593 UP000008144 UP000246464 UP000078492 UP000261640 UP000005205 UP000264820 UP000005207 UP000261620 UP000001038 UP000092460 UP000076502 UP000008672
Pfam
PF03690 UPF0160
Interpro
ProteinModelPortal
A0A2A4J1N7
A0A1E1WCX0
A0A2H1VTJ1
A0A0N0P9K5
A0A3S2TFW6
A0A212F6A6
+ More
A0A2A4J316 A0A194RKW5 D6WP32 A0A1E1WQN8 A0A182P710 A0A0T6BA06 A0A182U8B0 Q7QC19 U5EVU0 A0A182LFA3 A0A1Y9J005 A0A182VX73 A0A182HT64 A0A182UQL5 A0A182K7C7 N6TNI3 A0A1W4X8Y9 A0A182T8M8 A0A1W4XJF3 A0A1W4XJ63 T1EA83 A0A182YKG3 Q172K4 A0A182J6G6 B0XKE1 A0A182MEA5 A0A182QRS7 A0A182NA65 A0A336LPI0 A0A2M4APH7 E0VKY8 W5JK47 Q172K5 A0A1J1IHR0 A0A182H0R0 A0A1Q3F1L4 A0A182H139 A0A023ERL7 A0A084VSD6 A0A1B0D999 A0A1Y1MX31 A0A1Y1MU12 A0A2T7PJ00 A0A0K8TR04 A0A067RCM3 A0A1L8E032 R4G836 A0A0P7ZDL0 K7J738 A0A2S2Q1Y5 W5ME83 A0A2J7PQK2 A0A1W4ZAJ3 A0A2J7PQG3 A0A1B6DLN8 A0A3B3R8M2 A0A195D5R8 A9UV55 A0A0A1WJ22 V4A4D9 R7V123 A0A0A1WWD0 Q4RX01 H3DBH6 A0A170XUD1 A0A3Q3MIC2 A0A195AZH1 C3XWS9 A0A1W2WJA3 A0A1W2WR78 H2ZNV1 E9I8N9 A0A195FWN8 F4WD91 A7S559 F6ZCS4 C3XWP2 A0A2H8TFE1 A0A2U9BF80 W8C9P3 A0A195E241 A0A3Q3LYS5 A0A2S2N8G4 A0A158NHN3 A0A0V0G881 A0A3Q3ECQ1 I3JC73 A0A3Q4B731 H2LKA5 A0A1B0BSA6 A0A154PA46 H3B2C3 A0A224XSQ5
A0A2A4J316 A0A194RKW5 D6WP32 A0A1E1WQN8 A0A182P710 A0A0T6BA06 A0A182U8B0 Q7QC19 U5EVU0 A0A182LFA3 A0A1Y9J005 A0A182VX73 A0A182HT64 A0A182UQL5 A0A182K7C7 N6TNI3 A0A1W4X8Y9 A0A182T8M8 A0A1W4XJF3 A0A1W4XJ63 T1EA83 A0A182YKG3 Q172K4 A0A182J6G6 B0XKE1 A0A182MEA5 A0A182QRS7 A0A182NA65 A0A336LPI0 A0A2M4APH7 E0VKY8 W5JK47 Q172K5 A0A1J1IHR0 A0A182H0R0 A0A1Q3F1L4 A0A182H139 A0A023ERL7 A0A084VSD6 A0A1B0D999 A0A1Y1MX31 A0A1Y1MU12 A0A2T7PJ00 A0A0K8TR04 A0A067RCM3 A0A1L8E032 R4G836 A0A0P7ZDL0 K7J738 A0A2S2Q1Y5 W5ME83 A0A2J7PQK2 A0A1W4ZAJ3 A0A2J7PQG3 A0A1B6DLN8 A0A3B3R8M2 A0A195D5R8 A9UV55 A0A0A1WJ22 V4A4D9 R7V123 A0A0A1WWD0 Q4RX01 H3DBH6 A0A170XUD1 A0A3Q3MIC2 A0A195AZH1 C3XWS9 A0A1W2WJA3 A0A1W2WR78 H2ZNV1 E9I8N9 A0A195FWN8 F4WD91 A7S559 F6ZCS4 C3XWP2 A0A2H8TFE1 A0A2U9BF80 W8C9P3 A0A195E241 A0A3Q3LYS5 A0A2S2N8G4 A0A158NHN3 A0A0V0G881 A0A3Q3ECQ1 I3JC73 A0A3Q4B731 H2LKA5 A0A1B0BSA6 A0A154PA46 H3B2C3 A0A224XSQ5
Ontologies
PANTHER
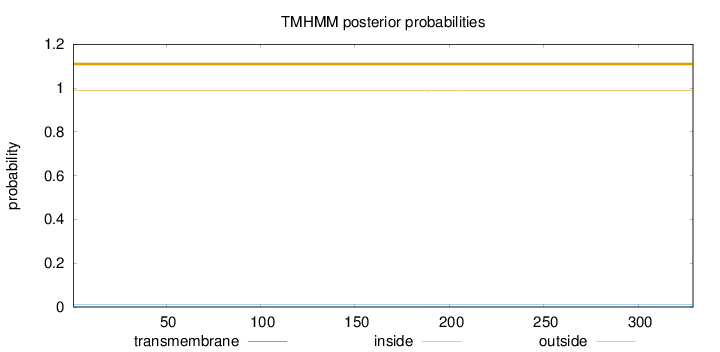
Topology
Length:
329
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01279
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01240
outside
1 - 329
Population Genetic Test Statistics
Pi
241.92128
Theta
166.892431
Tajima's D
1.875907
CLR
0.790788
CSRT
0.864456777161142
Interpretation
Uncertain
