Gene
KWMTBOMO04111
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.747 Nuclear Reliability : 1.731
Sequence
CDS
ATGGACGCTGTATTTGCGGAATTTCTCCGGCTTCGCTACCCAAAACTCACTTCTGAGTTTCAGGCCTTCAAGGCCGATCACACCGCGAGCCCTCTCGTGGACTCCGCTGAGCTTGCTGCTCCTGCGTCGCCTGTACCTGCGTGCAAAGCTTTTGTATCGAGCACCGCCGCCTCCGTCGTGCCTGCCGTTCCCGCGTCGCCTATACTGGCGAGTAAAACTGCTGCGTCGTCCGCCGTGGTCACCGTTGCGCCCTCCAAAACACCTACTCGTAGGTCACCTGCGCCCGCCTCCTCGTCCTCCGACTCTGACTCGGAGATGGAGGTCGACCTCGCCCCCGTCTCATCGACGGATGGATTCACACTGGTGCAAAAGGGTAAGAAGCGTGCCGCGGAGTCTCGAGCTCCCGCGGCCGCTAAAATTAGCAAAGCCGCGAACGCGTCGCGCCCCCGCCCTCAGACTCCCGTTGCGCCTCCAGCCCGTGCCACTCCGTCGCCGCGTCCGGTGGCACAAAATAAAACCCAGACCCCTCCCCCGGTAATCCTTCAAGAGAAGGCAGCTTGGGAACGAGTTGCCCTGGCCCTTAAGGCCAAAAATATCAATTTTACGAATGCCCGTAACCTCGCGAACGGCATTCAAATTAAGGTTCAAACACCCGACGACCATAGGGCCCTCTCTTCTTACCTCCGTAAGGAGCGTATAAGTTTCCACACGTATACGCTCCAGGAGGAGCGCGAACTCCGTGCCGTTATACGTGGCATCCCTAAAGAGTTGGATGCCGAGCTCGTCAAGGCCGACCTTCTCGAACAAGGCCTACCGGTTAACTCCGTACACCGCATGCACACAGGCCGCGGAAGGGAGCCATATAATATGGTTCTCGTCGCCCTCCAGCCTACCCCCGAGGCGGAGGACTTCCTTCACCGCTTCAAATTGTCCGACGATATTAGGGAACTCCTTAGAGCTAAGAACGCTTCGATACGCGCCTACGACAGGTATCCTACCGCGGAAAATCGTATTCGAATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGCCAGATGGTCTGATTTCTTAAAAGGACTCGCGCCCTCCCAAAGGTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTATGCCCCCACTCGCAGGCCCCTCAGGCCGACTCACGGCGTTCGATGATGACGAAAAAACAGAGCTGTTGGCCGATACATTGCAAACCCAGTGTACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCCTCGGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCCAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGATATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATCAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTCATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAACGGTTTGATTTTCAAACTATTCAACATGGGTGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTTACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTTAACGATATTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCCAGTAGAAACAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTATTTCAGAGGGGAAGCTCCACACGGATTTCCTCCCGTATTAGGAGGAGGAATCTCACACCCCCGATTACTCTCTTTAGTCAATCCATAACCTGGGCCAGGAAGGTCAAGTACCTGGGCGTTACCCTGGATGCATCGATGACATTCCGCCCGCATATAAAATCAGTCCGTGACCGTGCCGCGTTTATTCTCGGTAGACTCTACCCCATGATCTGTAAGCAGAGTAAAATGTCCCTTCGGAACAAGGTGACACTTTACAAAACTTGCATAAGGCCCGTCATGACTTACGCGAGTGTGGTGTTCGCTCACGCGGCCCGCACACACATAGACACCCTTCAATCTCTACAATCCCGCTTTTGCAGGTTAGCCGTCGGAGCTCCGTGGTTCGTGAGGAACGTTGACCTACACGACGACCTGGGCCTCGAATCTATACAGAAATACATGAAGTCAGCGTCGGAACGGTACTTCGATAAGGCTATGCGTCATGATAATCGCCTTATCGTAGCCGCCGCTGACTACTCCCCGAATCCTGATCATGCAGGAGCCAGTCACCGTCGACGCCCTAGACACGTCCTTACGGACCCATCAGATCCAATAACCTTTGCACTAGACGCCTTCAGCTCTAGGAGCAGGCTTAGGGACCTCGATTTCACCAAGACTACACTATAG
Protein
MDAVFAEFLRLRYPKLTSEFQAFKADHTASPLVDSAELAAPASPVPACKAFVSSTAASVVPAVPASPILASKTAASSAVVTVAPSKTPTRRSPAPASSSSDSDSEMEVDLAPVSSTDGFTLVQKGKKRAAESRAPAAAKISKAANASRPRPQTPVAPPARATPSPRPVAQNKTQTPPPVILQEKAAWERVALALKAKNINFTNARNLANGIQIKVQTPDDHRALSSYLRKERISFHTYTLQEERELRAVIRGIPKELDAELVKADLLEQGLPVNSVHRMHTGRGREPYNMVLVALQPTPEAEDFLHRFKLSDDIRELLRAKNASIRAYDRYPTAENRIRMRALQRDVKSRIAEVRDARWSDFLKGLAPSQRSYYRLARTLKSDTVVTMPPLAGPSGRLTAFDDDEKTELLADTLQTQCTPSTQSVDPVHVELVDSEVERRASLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADDIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDIPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQRGSSTRISSRIRRRNLTPPITLFSQSITWARKVKYLGVTLDASMTFRPHIKSVRDRAAFILGRLYPMICKQSKMSLRNKVTLYKTCIRPVMTYASVVFAHAARTHIDTLQSLQSRFCRLAVGAPWFVRNVDLHDDLGLESIQKYMKSASERYFDKAMRHDNRLIVAAADYSPNPDHAGASHRRRPRHVLTDPSDPITFALDAFSSRSRLRDLDFTKTTL
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
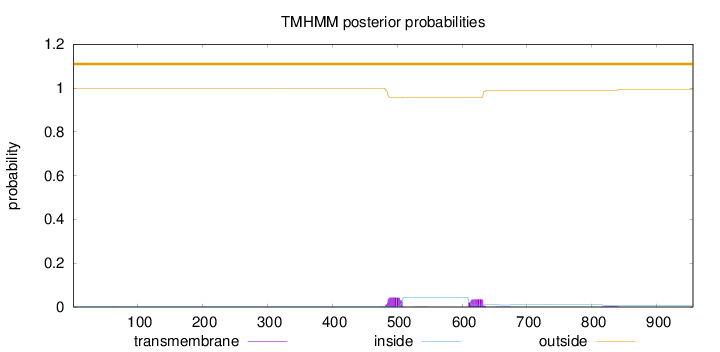
Topology
Length:
956
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.81536
Exp number, first 60 AAs:
0.0009
Total prob of N-in:
0.00110
outside
1 - 956
Population Genetic Test Statistics
Pi
0.974992
Theta
0.336228
Tajima's D
2.256684
CLR
0.674405
CSRT
0.999950002499875
Interpretation
Uncertain
