Pre Gene Modal
BGIBMGA008612
Annotation
PREDICTED:_phosphatidylinositol_4-kinase_type_2-beta_isoform_X1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.557
Sequence
CDS
ATGAGTACGGAAGAGACTCTGTTGAATCTCGATTTCGACAAACCAAACACAACAACAGCACCGCCCGATGACGGAGTGGTCTCGTTATCCAGTAGTTCAGAAGAGATTTGTTTAGTGCCAGAGGTCGAGTTCGAGGGCAATCTGGATTCTCCAGGAGTGAACCGCGAGTCGCAGCCGCTGCTTGGCGGCCGCGGCGACTTCGAAGTATCCTTCAATTACTTCCCAGATGACCCCCAGTTCAGCGAGCTGGTGCTACAGGCGGAGACGGCCATTGATAACGGCATATTCCCGGAGAGGATATATCAAGGATCCAGCGGGTCGTACTTCGTAAAAAACGCGGGCGGGAAAATCATCGGCGTTTTCAAGCCGAAAGACGAAGAGCCCTACGGTCGTTTGAACCCCAAATGGATGAAATGGATGCACAAGCTTTGCTGCCCGTGTTGCTTCGGCCGCTCCTGTCTGATCCCGAACCAGGGGTACTTGTCCGAGGCCGGTGCGAGCCTCGTAGATTCCAAAATAGGACTTAAAGTCGTGCCTAAGACTAAGGTAGTAAAATTAGTATCGGAAACGTTTAACTATCTGCGCATAGACCGAGAGAGGTCGCGTCTGAAGCGGGCCATCACCGAGCAGTTCCCGAACTTGCGATTCAACAGGATGGGGCTGCCTCCTAAGAAAGGCTCATTCCAGTTATTCGTGGAGGGCTACAAGGACGCGGACTATTGGCTCAGGAGGTTCGAGCAGGAGCCCCCGCCGCCGCACGTCATGAAGAAGTTCCAGCTGCAGTTCGAGCGGCTCGTGGTGCTGGACTACATCATCCGGAACACGGACCGCGGCAACGACAACTGGCTCATCAAGTACGAGGCGCCGCCCGGCCCGCGCGCCGACGCCGACCTGCGGGACCCCTCGGAGTGGTCGGGCGGGTGGGAGGTGCGCATCGCGGCCATCGACAACGGGCTCGCGTTCCCGTTCAAGCACCCGGACTCGTGGCGCGCCTACCCCTACCACTGGGCCTGGCTGCCGCACGCCAAGCTGCCCTTCAGCGACGAGACCAAGCAACTAGTACTTCCGCTGTTATCCGACATGAACTTCGTTCAGGAACTGTGCGACGAACTACATTTGTTGTTTAAACAAGACAAGGGCTTCGACAAGGGTCTGTTCGAGCGGCAGATGTCGGTGATGCGCGGGCAGGTGCTCAACCTCACGCAGGCCCTCAAGGACGGCAAGAGTCCCGTGCAGCTGGTGCAGATGCCGTGCGTGCTCGTCGAGAGGTAA
Protein
MSTEETLLNLDFDKPNTTTAPPDDGVVSLSSSSEEICLVPEVEFEGNLDSPGVNRESQPLLGGRGDFEVSFNYFPDDPQFSELVLQAETAIDNGIFPERIYQGSSGSYFVKNAGGKIIGVFKPKDEEPYGRLNPKWMKWMHKLCCPCCFGRSCLIPNQGYLSEAGASLVDSKIGLKVVPKTKVVKLVSETFNYLRIDRERSRLKRAITEQFPNLRFNRMGLPPKKGSFQLFVEGYKDADYWLRRFEQEPPPPHVMKKFQLQFERLVVLDYIIRNTDRGNDNWLIKYEAPPGPRADADLRDPSEWSGGWEVRIAAIDNGLAFPFKHPDSWRAYPYHWAWLPHAKLPFSDETKQLVLPLLSDMNFVQELCDELHLLFKQDKGFDKGLFERQMSVMRGQVLNLTQALKDGKSPVQLVQMPCVLVER
Summary
Uniprot
A0A2A4K5W4
A0A194RPK8
A0A194Q092
S4P987
A0A2H1W6D8
N6UGL3
+ More
D7EK02 J3JV24 A0A1W4XV42 A0A1Y1KAC9 A0A1L8DM19 A0A1L8DMB6 A0A1L8DMA4 A0A2J7QLR6 A0A0K8TRF9 A0A2J7QLT2 A0A232FC45 A0A1S4F7E9 K7IWW3 Q17CZ9 B0X9F6 A0A1Q3FAG8 A0A1Q3F9W4 A0A1Q3FB72 A0A2J7QLT7 A0A067RIZ9 A0A154PFB6 A0A0L7QKP6 A0A182MMS0 A0A084W4Y7 A0A182QQL3 A0A0M9A989 A0A310SAW4 A0A1B6C7D0 A0A088AFG2 A0A0P4WJ62 A0A2A3ECE7 E2AP86 A0A0J7KTQ5 F4WR56 E9IHK1 A0A195BBR7 A0A158NU78 A0A151IQF1 A0A0P5YTZ8 A0A151J3B1 A0A0P6HC93 A0A195FLJ2 E2BNI7 A0A0P5QEI9 A0A0P5VHB4 A0A026W960 A0A1B6HUU1 A0A2S2P431 A0A2H8TVM4 A0A2S2QUD9 J9JV58 A0A336LJ27 A0A336K067 A0A151WEX8 A0A182PFG3 A0A0P4WFC1 A0A182XVL7 A0A182W2V1 A0A1D2MGN8 A0A224Z506 A0A131YJ29 A0A2T7P7M5 L7MJN5 A0A0P4WC19 A0A0N7ZD32 A0A131XKQ4 T1IIS3 G3MGJ6 A0A0K8RC37 A0A023G4H2 A0A1E1XCC9 A0A023FMD1 A0A226DFN8 A0A0P5HVZ8 A0A3R7P6C5 A0A1S3IMP5 A0A0C9SEA1 A0A2C9K855 A0A2R5LL98 A0A0N8C396 A0A293LY81 A0A3B3WLN2 M3ZTP1 A0A3B3WLD3 A0A1S3INJ5 A0A087Y9X6 A0A3B3TW46 A0A0S7L8E5 A0A1B6J0U9 E0VSG9 A0A3Q2FVK5 A0A146MZH8 A0A3Q1JBZ4 A0A3P9MTN5
D7EK02 J3JV24 A0A1W4XV42 A0A1Y1KAC9 A0A1L8DM19 A0A1L8DMB6 A0A1L8DMA4 A0A2J7QLR6 A0A0K8TRF9 A0A2J7QLT2 A0A232FC45 A0A1S4F7E9 K7IWW3 Q17CZ9 B0X9F6 A0A1Q3FAG8 A0A1Q3F9W4 A0A1Q3FB72 A0A2J7QLT7 A0A067RIZ9 A0A154PFB6 A0A0L7QKP6 A0A182MMS0 A0A084W4Y7 A0A182QQL3 A0A0M9A989 A0A310SAW4 A0A1B6C7D0 A0A088AFG2 A0A0P4WJ62 A0A2A3ECE7 E2AP86 A0A0J7KTQ5 F4WR56 E9IHK1 A0A195BBR7 A0A158NU78 A0A151IQF1 A0A0P5YTZ8 A0A151J3B1 A0A0P6HC93 A0A195FLJ2 E2BNI7 A0A0P5QEI9 A0A0P5VHB4 A0A026W960 A0A1B6HUU1 A0A2S2P431 A0A2H8TVM4 A0A2S2QUD9 J9JV58 A0A336LJ27 A0A336K067 A0A151WEX8 A0A182PFG3 A0A0P4WFC1 A0A182XVL7 A0A182W2V1 A0A1D2MGN8 A0A224Z506 A0A131YJ29 A0A2T7P7M5 L7MJN5 A0A0P4WC19 A0A0N7ZD32 A0A131XKQ4 T1IIS3 G3MGJ6 A0A0K8RC37 A0A023G4H2 A0A1E1XCC9 A0A023FMD1 A0A226DFN8 A0A0P5HVZ8 A0A3R7P6C5 A0A1S3IMP5 A0A0C9SEA1 A0A2C9K855 A0A2R5LL98 A0A0N8C396 A0A293LY81 A0A3B3WLN2 M3ZTP1 A0A3B3WLD3 A0A1S3INJ5 A0A087Y9X6 A0A3B3TW46 A0A0S7L8E5 A0A1B6J0U9 E0VSG9 A0A3Q2FVK5 A0A146MZH8 A0A3Q1JBZ4 A0A3P9MTN5
Pubmed
EMBL
NWSH01000139
PCG79163.1
KQ460075
KPJ17956.1
KQ459582
KPI98718.1
+ More
GAIX01005616 JAA86944.1 ODYU01006604 SOQ48593.1 APGK01021327 APGK01021328 KB740314 KB631743 ENN80890.1 ERL85729.1 DS497724 EFA12949.1 BT127090 AEE62052.1 GEZM01092329 JAV56595.1 GFDF01006582 JAV07502.1 GFDF01006580 JAV07504.1 GFDF01006579 JAV07505.1 NEVH01013241 PNF29535.1 GDAI01000651 JAI16952.1 PNF29536.1 NNAY01000459 OXU28245.1 AAZX01006908 CH477301 EAT44260.1 DS232534 EDS43125.1 GFDL01010533 JAV24512.1 GFDL01010685 JAV24360.1 GFDL01010297 JAV24748.1 PNF29537.1 KK852478 KDR23003.1 KQ434892 KZC10553.1 KQ414940 KOC59182.1 AXCM01000405 AXCM01000406 ATLV01020424 KE525302 KFB45281.1 AXCN02000061 KQ435710 KOX79807.1 KQ777939 OAD52034.1 GEDC01027905 JAS09393.1 GDRN01054073 JAI66121.1 KZ288287 PBC29398.1 GL441481 EFN64759.1 LBMM01003198 KMQ93817.1 GL888284 EGI63283.1 GL763289 EFZ19898.1 KQ976532 KYM81639.1 ADTU01026286 KQ976780 KYN08377.1 GDIP01053469 GDIQ01084709 JAM50246.1 JAN10028.1 KQ980298 KYN16842.1 GDIQ01021291 JAN73446.1 KQ981512 KYN40859.1 GL449414 EFN82766.1 GDIQ01121895 JAL29831.1 GDIP01145992 GDIQ01210509 GDIQ01145341 GDIP01099937 LRGB01000190 JAK41216.1 JAM03778.1 KZS20443.1 KK107323 QOIP01000002 EZA52607.1 RLU25557.1 GECU01029252 GECU01023962 JAS78454.1 JAS83744.1 GGMR01011483 MBY24102.1 GFXV01006548 MBW18353.1 GGMS01012166 MBY81369.1 ABLF02034412 UFQS01000027 UFQT01000027 SSW97758.1 SSX18144.1 SSW97757.1 SSX18143.1 KQ983238 KYQ46356.1 GDRN01054072 JAI66122.1 LJIJ01001315 ODM92158.1 GFPF01010547 MAA21693.1 GEDV01010626 JAP77931.1 PZQS01000005 PVD29409.1 GACK01000859 JAA64175.1 GDRN01054071 JAI66123.1 GDRN01054074 JAI66120.1 GEFH01001863 JAP66718.1 JH430212 JO840997 AEO32614.1 GADI01005113 JAA68695.1 GBBM01006694 JAC28724.1 GFAC01002284 JAT96904.1 GBBK01002497 JAC21985.1 LNIX01000020 OXA43950.1 GDIQ01228047 JAK23678.1 QCYY01003717 ROT62363.1 GBZX01001337 JAG91403.1 GGLE01005971 MBY10097.1 GDIQ01119091 JAL32635.1 GFWV01007080 MAA31810.1 AYCK01001663 GBYX01181294 JAO85799.1 GECU01014891 JAS92815.1 DS235751 EEB16325.1 GCES01162156 JAQ24166.1
GAIX01005616 JAA86944.1 ODYU01006604 SOQ48593.1 APGK01021327 APGK01021328 KB740314 KB631743 ENN80890.1 ERL85729.1 DS497724 EFA12949.1 BT127090 AEE62052.1 GEZM01092329 JAV56595.1 GFDF01006582 JAV07502.1 GFDF01006580 JAV07504.1 GFDF01006579 JAV07505.1 NEVH01013241 PNF29535.1 GDAI01000651 JAI16952.1 PNF29536.1 NNAY01000459 OXU28245.1 AAZX01006908 CH477301 EAT44260.1 DS232534 EDS43125.1 GFDL01010533 JAV24512.1 GFDL01010685 JAV24360.1 GFDL01010297 JAV24748.1 PNF29537.1 KK852478 KDR23003.1 KQ434892 KZC10553.1 KQ414940 KOC59182.1 AXCM01000405 AXCM01000406 ATLV01020424 KE525302 KFB45281.1 AXCN02000061 KQ435710 KOX79807.1 KQ777939 OAD52034.1 GEDC01027905 JAS09393.1 GDRN01054073 JAI66121.1 KZ288287 PBC29398.1 GL441481 EFN64759.1 LBMM01003198 KMQ93817.1 GL888284 EGI63283.1 GL763289 EFZ19898.1 KQ976532 KYM81639.1 ADTU01026286 KQ976780 KYN08377.1 GDIP01053469 GDIQ01084709 JAM50246.1 JAN10028.1 KQ980298 KYN16842.1 GDIQ01021291 JAN73446.1 KQ981512 KYN40859.1 GL449414 EFN82766.1 GDIQ01121895 JAL29831.1 GDIP01145992 GDIQ01210509 GDIQ01145341 GDIP01099937 LRGB01000190 JAK41216.1 JAM03778.1 KZS20443.1 KK107323 QOIP01000002 EZA52607.1 RLU25557.1 GECU01029252 GECU01023962 JAS78454.1 JAS83744.1 GGMR01011483 MBY24102.1 GFXV01006548 MBW18353.1 GGMS01012166 MBY81369.1 ABLF02034412 UFQS01000027 UFQT01000027 SSW97758.1 SSX18144.1 SSW97757.1 SSX18143.1 KQ983238 KYQ46356.1 GDRN01054072 JAI66122.1 LJIJ01001315 ODM92158.1 GFPF01010547 MAA21693.1 GEDV01010626 JAP77931.1 PZQS01000005 PVD29409.1 GACK01000859 JAA64175.1 GDRN01054071 JAI66123.1 GDRN01054074 JAI66120.1 GEFH01001863 JAP66718.1 JH430212 JO840997 AEO32614.1 GADI01005113 JAA68695.1 GBBM01006694 JAC28724.1 GFAC01002284 JAT96904.1 GBBK01002497 JAC21985.1 LNIX01000020 OXA43950.1 GDIQ01228047 JAK23678.1 QCYY01003717 ROT62363.1 GBZX01001337 JAG91403.1 GGLE01005971 MBY10097.1 GDIQ01119091 JAL32635.1 GFWV01007080 MAA31810.1 AYCK01001663 GBYX01181294 JAO85799.1 GECU01014891 JAS92815.1 DS235751 EEB16325.1 GCES01162156 JAQ24166.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000019118
UP000030742
UP000007266
+ More
UP000192223 UP000235965 UP000215335 UP000002358 UP000008820 UP000002320 UP000027135 UP000076502 UP000053825 UP000075883 UP000030765 UP000075886 UP000053105 UP000005203 UP000242457 UP000000311 UP000036403 UP000007755 UP000078540 UP000005205 UP000078542 UP000078492 UP000078541 UP000008237 UP000076858 UP000053097 UP000279307 UP000007819 UP000075809 UP000075885 UP000076408 UP000075920 UP000094527 UP000245119 UP000198287 UP000283509 UP000085678 UP000076420 UP000261480 UP000002852 UP000028760 UP000261500 UP000009046 UP000265020 UP000265040 UP000242638
UP000192223 UP000235965 UP000215335 UP000002358 UP000008820 UP000002320 UP000027135 UP000076502 UP000053825 UP000075883 UP000030765 UP000075886 UP000053105 UP000005203 UP000242457 UP000000311 UP000036403 UP000007755 UP000078540 UP000005205 UP000078542 UP000078492 UP000078541 UP000008237 UP000076858 UP000053097 UP000279307 UP000007819 UP000075809 UP000075885 UP000076408 UP000075920 UP000094527 UP000245119 UP000198287 UP000283509 UP000085678 UP000076420 UP000261480 UP000002852 UP000028760 UP000261500 UP000009046 UP000265020 UP000265040 UP000242638
Interpro
CDD
ProteinModelPortal
A0A2A4K5W4
A0A194RPK8
A0A194Q092
S4P987
A0A2H1W6D8
N6UGL3
+ More
D7EK02 J3JV24 A0A1W4XV42 A0A1Y1KAC9 A0A1L8DM19 A0A1L8DMB6 A0A1L8DMA4 A0A2J7QLR6 A0A0K8TRF9 A0A2J7QLT2 A0A232FC45 A0A1S4F7E9 K7IWW3 Q17CZ9 B0X9F6 A0A1Q3FAG8 A0A1Q3F9W4 A0A1Q3FB72 A0A2J7QLT7 A0A067RIZ9 A0A154PFB6 A0A0L7QKP6 A0A182MMS0 A0A084W4Y7 A0A182QQL3 A0A0M9A989 A0A310SAW4 A0A1B6C7D0 A0A088AFG2 A0A0P4WJ62 A0A2A3ECE7 E2AP86 A0A0J7KTQ5 F4WR56 E9IHK1 A0A195BBR7 A0A158NU78 A0A151IQF1 A0A0P5YTZ8 A0A151J3B1 A0A0P6HC93 A0A195FLJ2 E2BNI7 A0A0P5QEI9 A0A0P5VHB4 A0A026W960 A0A1B6HUU1 A0A2S2P431 A0A2H8TVM4 A0A2S2QUD9 J9JV58 A0A336LJ27 A0A336K067 A0A151WEX8 A0A182PFG3 A0A0P4WFC1 A0A182XVL7 A0A182W2V1 A0A1D2MGN8 A0A224Z506 A0A131YJ29 A0A2T7P7M5 L7MJN5 A0A0P4WC19 A0A0N7ZD32 A0A131XKQ4 T1IIS3 G3MGJ6 A0A0K8RC37 A0A023G4H2 A0A1E1XCC9 A0A023FMD1 A0A226DFN8 A0A0P5HVZ8 A0A3R7P6C5 A0A1S3IMP5 A0A0C9SEA1 A0A2C9K855 A0A2R5LL98 A0A0N8C396 A0A293LY81 A0A3B3WLN2 M3ZTP1 A0A3B3WLD3 A0A1S3INJ5 A0A087Y9X6 A0A3B3TW46 A0A0S7L8E5 A0A1B6J0U9 E0VSG9 A0A3Q2FVK5 A0A146MZH8 A0A3Q1JBZ4 A0A3P9MTN5
D7EK02 J3JV24 A0A1W4XV42 A0A1Y1KAC9 A0A1L8DM19 A0A1L8DMB6 A0A1L8DMA4 A0A2J7QLR6 A0A0K8TRF9 A0A2J7QLT2 A0A232FC45 A0A1S4F7E9 K7IWW3 Q17CZ9 B0X9F6 A0A1Q3FAG8 A0A1Q3F9W4 A0A1Q3FB72 A0A2J7QLT7 A0A067RIZ9 A0A154PFB6 A0A0L7QKP6 A0A182MMS0 A0A084W4Y7 A0A182QQL3 A0A0M9A989 A0A310SAW4 A0A1B6C7D0 A0A088AFG2 A0A0P4WJ62 A0A2A3ECE7 E2AP86 A0A0J7KTQ5 F4WR56 E9IHK1 A0A195BBR7 A0A158NU78 A0A151IQF1 A0A0P5YTZ8 A0A151J3B1 A0A0P6HC93 A0A195FLJ2 E2BNI7 A0A0P5QEI9 A0A0P5VHB4 A0A026W960 A0A1B6HUU1 A0A2S2P431 A0A2H8TVM4 A0A2S2QUD9 J9JV58 A0A336LJ27 A0A336K067 A0A151WEX8 A0A182PFG3 A0A0P4WFC1 A0A182XVL7 A0A182W2V1 A0A1D2MGN8 A0A224Z506 A0A131YJ29 A0A2T7P7M5 L7MJN5 A0A0P4WC19 A0A0N7ZD32 A0A131XKQ4 T1IIS3 G3MGJ6 A0A0K8RC37 A0A023G4H2 A0A1E1XCC9 A0A023FMD1 A0A226DFN8 A0A0P5HVZ8 A0A3R7P6C5 A0A1S3IMP5 A0A0C9SEA1 A0A2C9K855 A0A2R5LL98 A0A0N8C396 A0A293LY81 A0A3B3WLN2 M3ZTP1 A0A3B3WLD3 A0A1S3INJ5 A0A087Y9X6 A0A3B3TW46 A0A0S7L8E5 A0A1B6J0U9 E0VSG9 A0A3Q2FVK5 A0A146MZH8 A0A3Q1JBZ4 A0A3P9MTN5
PDB
4HNE
E-value=3.54862e-127,
Score=1165
Ontologies
PATHWAY
GO
PANTHER
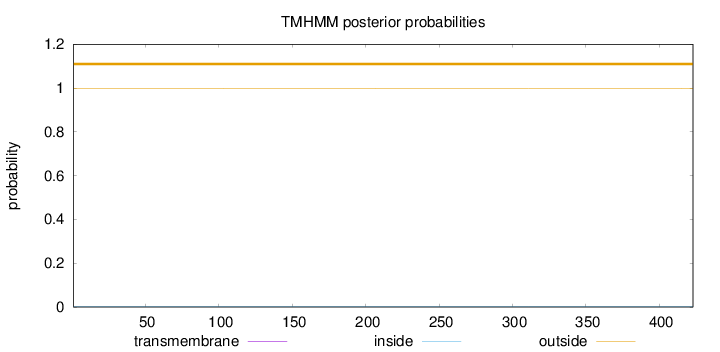
Topology
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00206
outside
1 - 423
Population Genetic Test Statistics
Pi
256.346928
Theta
147.116929
Tajima's D
2.413733
CLR
0.122767
CSRT
0.935903204839758
Interpretation
Uncertain
