Gene
KWMTBOMO04093
Pre Gene Modal
BGIBMGA008630
Annotation
PREDICTED:_protein_FAM160B1-like_[Amyelois_transitella]
Full name
Protein FAM160B1
Location in the cell
Nuclear Reliability : 1.926
Sequence
CDS
ATGCTTAGCAGATTCTCCAACGTGCTGCAAACTGCTGCCGATGTCTTGGCACCTCCAGCAACGAGACTCGAAGACTTTGAGTACCATTGGAAGATGATACTTAACTTCTATGAGCGGTTTGATGATAATTGTAAAATTCACATTGAAGATACTAAGATACCTCATCACCTGCATCAGATGCTACAGCTCATTGTTGATGAAGAAAAAGAACAACAAGGTGGCACGGTCGGTCCATGCCTGGAAGCCATAGTTCAGCGGTCCATATGCCTGTTAGATGCATTGACTGCGCTCGCGGCGGCCGACCGACCGCCCGGCGCGAGGTCATTGGCCCTAACAGCTGCTACGGCACTGCTGAGGAGAACTAAACAGCCTTGCGTCAATAGCGTTCATATTTATCGTCCCTTACAGCGAATGCTAGTCCAGTGCAACGAAAGTCCGGCGTCGCCTACAGAGAAAGAAGAAGTCGAATTGCTGCTGACTCTGTGCGGTTTGGTCCGCAAGGAGCCAGGCCTTGCCAACATCTTCACGACACCGTTCGTGGAAGACAAAACCTCGTTACTAACTATACCGGAGGACATTCAGAAACTGATACCACTGAAGAAAGTGCACACGCCAAGAAAGAATCCTTTGTTCGTGGATCTGACCCAGAATCAGCTACCGAACGTGTCCATAATCAGGACCTACGACAGGGACGATAGCAGTAACGGCGCCGAGTCCTTGGCAGATGACGTGTCCTGCAAGAGTCAGAAGACATTGTACGATGAAAATGACAAGTTTTTGTTAATAGATTTGCTGCTTTCGTATTTGGACAGCGCTGATAACCAAGTAGTACTGAGAGCCTGCGAGGGGATAATGATAGTGTCGTCGCTGCCGGAGGAGGACATCGCCAACCTGGTGACGAGCTGCAGCCCGGCCTGCGAGACCATCGTGGACAAGCTCGCGGCCAGGTACCGCCGCATCCCGCCCACCGTCGACCCCAACGACGTCGACCAGCTCAACATCACCTGGGCATACGTAGCACAAGACTTCGGTGACAAGTCTTATTCCAAGTTCCACGGCTACAGAGAACTGACCACATTTTTCTCGTGGCTGGATTATTGCGATACACTCATGAAGGAATGTCACCAGAGCATAGCGTCGCACCTGTCGAGGACGTTCCGACAGAACTTTCTGGAGGGCACGGCGGAGGCCGGGGTCACCGACCTGCGCCTCGCCGTGCTGGCCACCGCCGTGCTCAGCAAGTGTCTCAAGGTCGTCGACTCCACTGACCTCATTAACGAGTTCTCCAACTGGCTGGTGGGGGACGGCGAGGTGGCGGAGTGGCCTCTACTGCATACCCTCATCTCGAACTGCCTCTCGGACAACTACGACCTGGCATTAGAAACATTAAGGTTCTTTGAGAGTATAATAGAGAAGGGCACGGAGCACAGCATGGAGCGACTGGTACTGTCTCGGGTAGGGACGCGCGGGGAGAGGGAAACGAACAGGGAGGCAATGAAGCAGCTACAACACGAGGAACACGTGAGTGAGCAAATCAACGACATCCTCTCACACGAGGGGCTCGACCTCGGCGGACACGCGGGCGCCGCCGAAAACATACACACCATCGTCAACAGTTACCTGCTGTTGCTCCCGCGCGCCCTCCTCTCCGACCCGGTGGGCTCCGACTACGAACATTACATACACGACGCCCATAGACACTATCAGGTAAACTGTCTTATTTTTTTAATTGAAATCGATCGGTCAGGTCTGTGGTGA
Protein
MLSRFSNVLQTAADVLAPPATRLEDFEYHWKMILNFYERFDDNCKIHIEDTKIPHHLHQMLQLIVDEEKEQQGGTVGPCLEAIVQRSICLLDALTALAAADRPPGARSLALTAATALLRRTKQPCVNSVHIYRPLQRMLVQCNESPASPTEKEEVELLLTLCGLVRKEPGLANIFTTPFVEDKTSLLTIPEDIQKLIPLKKVHTPRKNPLFVDLTQNQLPNVSIIRTYDRDDSSNGAESLADDVSCKSQKTLYDENDKFLLIDLLLSYLDSADNQVVLRACEGIMIVSSLPEEDIANLVTSCSPACETIVDKLAARYRRIPPTVDPNDVDQLNITWAYVAQDFGDKSYSKFHGYRELTTFFSWLDYCDTLMKECHQSIASHLSRTFRQNFLEGTAEAGVTDLRLAVLATAVLSKCLKVVDSTDLINEFSNWLVGDGEVAEWPLLHTLISNCLSDNYDLALETLRFFESIIEKGTEHSMERLVLSRVGTRGERETNREAMKQLQHEEHVSEQINDILSHEGLDLGGHAGAAENIHTIVNSYLLLLPRALLSDPVGSDYEHYIHDAHRHYQVNCLIFLIEIDRSGLW
Summary
Similarity
Belongs to the UPF0518 family.
Keywords
Complete proteome
Reference proteome
Alternative splicing
Polymorphism
Feature
chain Protein FAM160B1
splice variant In isoform 2.
sequence variant In dbSNP:rs17853717.
splice variant In isoform 2.
sequence variant In dbSNP:rs17853717.
Uniprot
A0A2A4K4I2
A0A3S2N983
A0A2A4K5H0
A0A194RKV1
A0A194Q154
A0A212FCZ3
+ More
A0A0L7LDV4 A0A1S4FH46 A0A182G9A2 A0A182GKC3 A0A1Q3F5H6 A0A1W5AYZ1 T1IIV5 A0A3Q0R8J3 G3X6C8 A0A250YLH2 B8A6I3 A0A2K5TMH6 A0A2K6DJF0 A0A2K5R094 A4IG57 A0A3B4G762 A0A2K6DJE5 A0A2K5TMG4 A0JNG7 A0A3P8QVD8 A0A3P9BV65 A0A3B4ADK3 A0A2I3TI67 B8A6I4 A0A3Q3IUR7 A0A3Q2VKU2 H2Q2L9 W5N5T4 A0A2J8W6X1 U3E924 A0A0D9QXX7 A0A2K5NC97 U3DJ61 A0A2J8W6Z4 A0A096NTS7 Q5W0V3-2 A0A3B3RI00 A0A2K5NC82 H9ESE8 A0A3Q4HDP0 A0A3Q4HAJ1 Q5W0V3 A0A096NTS6 I3JV87 A0A2I2Z5Z5 A0A3P8XUA4 G3RZX4 G1PXG0 S7N730 A0A3Q7R7A0 A0A2K6F2V3 A0A3Q7RU55 A0A1D5RAL8 A0A2K6F2U5 A0A3Q7PRI2 A0A3P8ZRF7 A0A2U3WTX8 A0A3B3RJT4 A0A3P8ZRH5 A0A3Q3M3Q1 F6Q4S8 M3X3J7 A0A2Y9DHT5 A0A1D5P0U6 A0A3B4THC3 A0A3B4THI2 A0A0K8RWH5 A0A226MR75 A0A3Q1B5X8 H3B5R3 H0XE78 A0A3P8UGJ3 J3S8Q2 A0A1D5QKB6 A0A2Y9J4N9 A0A3L8SIV9 H0ZL31 A0A1W7RGN4 G3UMA0 A0A226PQH7 A0A0F8C3X6 F7BGL9 A0A2Y9I1S5 G3NHZ0 A0A218UUE7 A0A182S849 Q8CDM8-2 F6XT73 A0A151NUU2
A0A0L7LDV4 A0A1S4FH46 A0A182G9A2 A0A182GKC3 A0A1Q3F5H6 A0A1W5AYZ1 T1IIV5 A0A3Q0R8J3 G3X6C8 A0A250YLH2 B8A6I3 A0A2K5TMH6 A0A2K6DJF0 A0A2K5R094 A4IG57 A0A3B4G762 A0A2K6DJE5 A0A2K5TMG4 A0JNG7 A0A3P8QVD8 A0A3P9BV65 A0A3B4ADK3 A0A2I3TI67 B8A6I4 A0A3Q3IUR7 A0A3Q2VKU2 H2Q2L9 W5N5T4 A0A2J8W6X1 U3E924 A0A0D9QXX7 A0A2K5NC97 U3DJ61 A0A2J8W6Z4 A0A096NTS7 Q5W0V3-2 A0A3B3RI00 A0A2K5NC82 H9ESE8 A0A3Q4HDP0 A0A3Q4HAJ1 Q5W0V3 A0A096NTS6 I3JV87 A0A2I2Z5Z5 A0A3P8XUA4 G3RZX4 G1PXG0 S7N730 A0A3Q7R7A0 A0A2K6F2V3 A0A3Q7RU55 A0A1D5RAL8 A0A2K6F2U5 A0A3Q7PRI2 A0A3P8ZRF7 A0A2U3WTX8 A0A3B3RJT4 A0A3P8ZRH5 A0A3Q3M3Q1 F6Q4S8 M3X3J7 A0A2Y9DHT5 A0A1D5P0U6 A0A3B4THC3 A0A3B4THI2 A0A0K8RWH5 A0A226MR75 A0A3Q1B5X8 H3B5R3 H0XE78 A0A3P8UGJ3 J3S8Q2 A0A1D5QKB6 A0A2Y9J4N9 A0A3L8SIV9 H0ZL31 A0A1W7RGN4 G3UMA0 A0A226PQH7 A0A0F8C3X6 F7BGL9 A0A2Y9I1S5 G3NHZ0 A0A218UUE7 A0A182S849 Q8CDM8-2 F6XT73 A0A151NUU2
Pubmed
26354079
22118469
26227816
17510324
26483478
19393038
+ More
28087693 23594743 25186727 25463417 16136131 25243066 17974005 15164054 15489334 10997877 12168954 24275569 29240929 25319552 22398555 25069045 21993624 17431167 17975172 15592404 25727380 9215903 23025625 30282656 20360741 26358130 24621616 25835551 19892987 16141072 12693553 21183079 17495919 22293439
28087693 23594743 25186727 25463417 16136131 25243066 17974005 15164054 15489334 10997877 12168954 24275569 29240929 25319552 22398555 25069045 21993624 17431167 17975172 15592404 25727380 9215903 23025625 30282656 20360741 26358130 24621616 25835551 19892987 16141072 12693553 21183079 17495919 22293439
EMBL
NWSH01000139
PCG79165.1
RSAL01000183
RVE44882.1
PCG79164.1
KQ460075
+ More
KPJ17955.1 KQ459582 KPI98719.1 AGBW02009118 OWR51616.1 JTDY01001508 KOB73703.1 JXUM01008424 JXUM01008425 KQ560260 KXJ83499.1 JXUM01069911 JXUM01069912 KQ562578 KXJ75560.1 GFDL01012224 JAV22821.1 JH430212 GFFW01000422 JAV44366.1 BX957309 AQIA01072680 AQIA01072681 AQIA01072682 AQIA01072683 BC134937 AAI34938.1 BC126677 AACZ04051197 NBAG03000216 PNI81647.1 GABC01007295 GABF01000738 GABD01006519 GABE01007109 JAA04043.1 JAA21407.1 JAA26581.1 JAA37630.1 PNI81648.1 AHAT01023224 NDHI03003398 PNJ65519.1 GAMT01008509 GAMS01001015 JAB03352.1 JAB22121.1 AQIB01042413 GAMR01004008 GAMQ01004605 GAMP01005627 JAB29924.1 JAB37246.1 JAB47128.1 PNJ65520.1 AHZZ02036108 CR933692 AL137025 BC037207 AB046820 JU321551 JU472691 AFE65307.1 AFH29495.1 AERX01006524 CABD030076293 CABD030076294 CABD030076295 CABD030076296 AAPE02016454 KE163614 EPQ12864.1 JSUE03044376 JSUE03044377 JSUE03044378 AANG04000273 AADN05000306 GBKC01001280 JAG44790.1 MCFN01000520 OXB57762.1 AFYH01064672 AFYH01064673 AFYH01064674 AAQR03046808 JU174532 GBEX01001218 AFJ50058.1 JAI13342.1 QUSF01000019 RLW02300.1 ABQF01046917 GDAY02001126 JAV50301.1 AWGT02000026 OXB82336.1 KQ042804 KKF10427.1 MUZQ01000140 OWK56982.1 AK029835 AK046518 AK172373 BC023048 BC062949 BC079562 AK122530 AKHW03001922 KYO40534.1
KPJ17955.1 KQ459582 KPI98719.1 AGBW02009118 OWR51616.1 JTDY01001508 KOB73703.1 JXUM01008424 JXUM01008425 KQ560260 KXJ83499.1 JXUM01069911 JXUM01069912 KQ562578 KXJ75560.1 GFDL01012224 JAV22821.1 JH430212 GFFW01000422 JAV44366.1 BX957309 AQIA01072680 AQIA01072681 AQIA01072682 AQIA01072683 BC134937 AAI34938.1 BC126677 AACZ04051197 NBAG03000216 PNI81647.1 GABC01007295 GABF01000738 GABD01006519 GABE01007109 JAA04043.1 JAA21407.1 JAA26581.1 JAA37630.1 PNI81648.1 AHAT01023224 NDHI03003398 PNJ65519.1 GAMT01008509 GAMS01001015 JAB03352.1 JAB22121.1 AQIB01042413 GAMR01004008 GAMQ01004605 GAMP01005627 JAB29924.1 JAB37246.1 JAB47128.1 PNJ65520.1 AHZZ02036108 CR933692 AL137025 BC037207 AB046820 JU321551 JU472691 AFE65307.1 AFH29495.1 AERX01006524 CABD030076293 CABD030076294 CABD030076295 CABD030076296 AAPE02016454 KE163614 EPQ12864.1 JSUE03044376 JSUE03044377 JSUE03044378 AANG04000273 AADN05000306 GBKC01001280 JAG44790.1 MCFN01000520 OXB57762.1 AFYH01064672 AFYH01064673 AFYH01064674 AAQR03046808 JU174532 GBEX01001218 AFJ50058.1 JAI13342.1 QUSF01000019 RLW02300.1 ABQF01046917 GDAY02001126 JAV50301.1 AWGT02000026 OXB82336.1 KQ042804 KKF10427.1 MUZQ01000140 OWK56982.1 AK029835 AK046518 AK172373 BC023048 BC062949 BC079562 AK122530 AKHW03001922 KYO40534.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000069940 UP000249989 UP000192224 UP000261340 UP000009136 UP000000437 UP000233100 UP000233120 UP000233040 UP000261460 UP000265100 UP000265160 UP000261520 UP000002277 UP000261600 UP000264840 UP000018468 UP000008225 UP000029965 UP000233060 UP000028761 UP000005640 UP000261540 UP000261580 UP000005207 UP000001519 UP000265140 UP000001074 UP000286641 UP000233160 UP000286640 UP000006718 UP000245340 UP000261660 UP000011712 UP000248480 UP000000539 UP000261420 UP000198323 UP000257160 UP000008672 UP000005225 UP000265080 UP000248482 UP000276834 UP000007754 UP000007646 UP000198419 UP000002281 UP000248481 UP000007635 UP000197619 UP000075901 UP000000589 UP000002280 UP000050525
UP000069940 UP000249989 UP000192224 UP000261340 UP000009136 UP000000437 UP000233100 UP000233120 UP000233040 UP000261460 UP000265100 UP000265160 UP000261520 UP000002277 UP000261600 UP000264840 UP000018468 UP000008225 UP000029965 UP000233060 UP000028761 UP000005640 UP000261540 UP000261580 UP000005207 UP000001519 UP000265140 UP000001074 UP000286641 UP000233160 UP000286640 UP000006718 UP000245340 UP000261660 UP000011712 UP000248480 UP000000539 UP000261420 UP000198323 UP000257160 UP000008672 UP000005225 UP000265080 UP000248482 UP000276834 UP000007754 UP000007646 UP000198419 UP000002281 UP000248481 UP000007635 UP000197619 UP000075901 UP000000589 UP000002280 UP000050525
PRIDE
Pfam
PF10257 RAI16-like
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A2A4K4I2
A0A3S2N983
A0A2A4K5H0
A0A194RKV1
A0A194Q154
A0A212FCZ3
+ More
A0A0L7LDV4 A0A1S4FH46 A0A182G9A2 A0A182GKC3 A0A1Q3F5H6 A0A1W5AYZ1 T1IIV5 A0A3Q0R8J3 G3X6C8 A0A250YLH2 B8A6I3 A0A2K5TMH6 A0A2K6DJF0 A0A2K5R094 A4IG57 A0A3B4G762 A0A2K6DJE5 A0A2K5TMG4 A0JNG7 A0A3P8QVD8 A0A3P9BV65 A0A3B4ADK3 A0A2I3TI67 B8A6I4 A0A3Q3IUR7 A0A3Q2VKU2 H2Q2L9 W5N5T4 A0A2J8W6X1 U3E924 A0A0D9QXX7 A0A2K5NC97 U3DJ61 A0A2J8W6Z4 A0A096NTS7 Q5W0V3-2 A0A3B3RI00 A0A2K5NC82 H9ESE8 A0A3Q4HDP0 A0A3Q4HAJ1 Q5W0V3 A0A096NTS6 I3JV87 A0A2I2Z5Z5 A0A3P8XUA4 G3RZX4 G1PXG0 S7N730 A0A3Q7R7A0 A0A2K6F2V3 A0A3Q7RU55 A0A1D5RAL8 A0A2K6F2U5 A0A3Q7PRI2 A0A3P8ZRF7 A0A2U3WTX8 A0A3B3RJT4 A0A3P8ZRH5 A0A3Q3M3Q1 F6Q4S8 M3X3J7 A0A2Y9DHT5 A0A1D5P0U6 A0A3B4THC3 A0A3B4THI2 A0A0K8RWH5 A0A226MR75 A0A3Q1B5X8 H3B5R3 H0XE78 A0A3P8UGJ3 J3S8Q2 A0A1D5QKB6 A0A2Y9J4N9 A0A3L8SIV9 H0ZL31 A0A1W7RGN4 G3UMA0 A0A226PQH7 A0A0F8C3X6 F7BGL9 A0A2Y9I1S5 G3NHZ0 A0A218UUE7 A0A182S849 Q8CDM8-2 F6XT73 A0A151NUU2
A0A0L7LDV4 A0A1S4FH46 A0A182G9A2 A0A182GKC3 A0A1Q3F5H6 A0A1W5AYZ1 T1IIV5 A0A3Q0R8J3 G3X6C8 A0A250YLH2 B8A6I3 A0A2K5TMH6 A0A2K6DJF0 A0A2K5R094 A4IG57 A0A3B4G762 A0A2K6DJE5 A0A2K5TMG4 A0JNG7 A0A3P8QVD8 A0A3P9BV65 A0A3B4ADK3 A0A2I3TI67 B8A6I4 A0A3Q3IUR7 A0A3Q2VKU2 H2Q2L9 W5N5T4 A0A2J8W6X1 U3E924 A0A0D9QXX7 A0A2K5NC97 U3DJ61 A0A2J8W6Z4 A0A096NTS7 Q5W0V3-2 A0A3B3RI00 A0A2K5NC82 H9ESE8 A0A3Q4HDP0 A0A3Q4HAJ1 Q5W0V3 A0A096NTS6 I3JV87 A0A2I2Z5Z5 A0A3P8XUA4 G3RZX4 G1PXG0 S7N730 A0A3Q7R7A0 A0A2K6F2V3 A0A3Q7RU55 A0A1D5RAL8 A0A2K6F2U5 A0A3Q7PRI2 A0A3P8ZRF7 A0A2U3WTX8 A0A3B3RJT4 A0A3P8ZRH5 A0A3Q3M3Q1 F6Q4S8 M3X3J7 A0A2Y9DHT5 A0A1D5P0U6 A0A3B4THC3 A0A3B4THI2 A0A0K8RWH5 A0A226MR75 A0A3Q1B5X8 H3B5R3 H0XE78 A0A3P8UGJ3 J3S8Q2 A0A1D5QKB6 A0A2Y9J4N9 A0A3L8SIV9 H0ZL31 A0A1W7RGN4 G3UMA0 A0A226PQH7 A0A0F8C3X6 F7BGL9 A0A2Y9I1S5 G3NHZ0 A0A218UUE7 A0A182S849 Q8CDM8-2 F6XT73 A0A151NUU2
Ontologies
GO
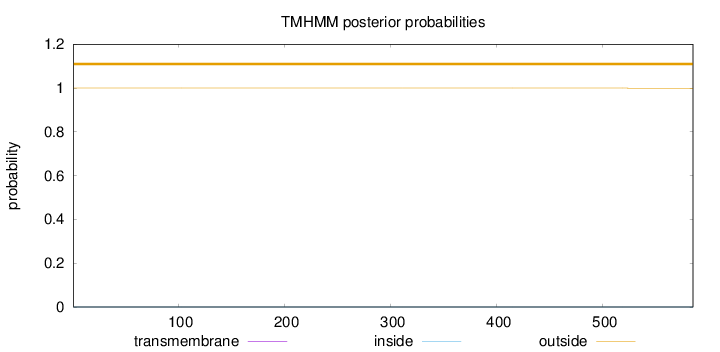
Topology
Length:
585
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00183
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00038
outside
1 - 585
Population Genetic Test Statistics
Pi
335.725647
Theta
181.164235
Tajima's D
1.701
CLR
0.173523
CSRT
0.827658617069147
Interpretation
Uncertain
