Gene
KWMTBOMO04082
Pre Gene Modal
BGIBMGA008626
Annotation
PREDICTED:_eukaryotic_translation_initiation_factor_2-alpha_kinase-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.994
Sequence
CDS
ATGTTGGACCAATTCGGTTTGATGAAGATTTTGTCTACTATTATTGCGTTATCGTACCTTGTTTGTTATCTTCAAGCAGAGGATGTTCCACTACAGCTACCGCACTGCAATCTTTCGAAGACTAAGGGATCAAGCTTAAACAATTATGTAATGGTGAGCACTTTGGATGGTCGCCTTACTGCATTTAATCTGGAAAATGGCACCAAAGCTTGGGTTGTTGAGACTCCACCATTGCTGTCCTCAAACTTGGCTAGCAGTGAGTTTAGTGGTGAAAATAGAGTGAAATTAGTGCCTTCACTAAGAGGCAGCTTGTACACTGTACATGGATCAAAGATCAAACGTTTACCATTCAATATACATCAGCTATTGTCTGAATCATTTACCGAGTCTAACAATCTCATCACTGCTGGCGGACATGACATACTACGGATAGGCATAGACACAAAAACAGGTCGTATTATATACAAATGTAACTCTAATGGATGCAACAATGAACAGCAAACAACAGAGACGGGAGGAGATGTACTGGTACTGACTCGGCAAGCCACTGCTATCAGAGGCCTAGACCCTAGAACAGGCATTGAGAAATGGAATTTTAGCACTGCCGCACATGAATTGCAGATAAGCCGTGGAGAGTGTATTGGGGATAGGACTGTAACCGAATCCCCAGTGAATATGCACGTATCAATGCATAACCAGCTGGTTACTGTCTTAAGCGGAGATCTAAATAAGCCGCTGTGGGAACATAAATTAGACGCGCCAATAACGGATGTATGGGAACTTTACGACGGTAACCTGCGAAATAGCCTGTGGCAGGATTCTCGCTCTAGCAATATTATGGGACAAATGCTCTACGTGGGTGATCATCAAGAACAGTTGTATATTTTGGATGAATTTGATCACTCTCAAAAGGACCGGAGCTTGATGGTGGTGCCATATCAATCGAGGAAGCGTTTCATCATTCTCGACAAAGAGAACGTGTACCTGCCTTTATCGGACAAGTCTACGTACATTCCGTTCCACGAGGCGGAGGACTCCAACGCCACCGAGGTCAGCGACAAGGTGAAGAAGGTGCGGTCGTGGTTATTCGACATCCTCACGTGCTTCACGCTGGCTTTGGCGTTAGTCTTGTTAATATTCCCTTTCTATGCGAGGATCCGAGTTATGATAACGAACGTCCCGCAAACTACGGTCAATGTAGTCTTCAACGAGGACATACCATCGGAGAAGGAGCTCGAAGCGAAAAGACCATCGAATCCCGAGTATCCCGGACGATATGAGAAGGACTTCACGACCGTGAGGCGCCTGGGGAGGGGCGGCTTCGGGGTCGTCTTCGAGGCGAGGAACAACATCGACAACTGTTCGTATGCCGTCAAGAGGATCACTCTGCCGAGGAAGAAGGAGCGGCACGAGCGCGTGCTGCGGGAGGCGCGCGCGCTCGCCAACCTCGAGCATCCGCACATTGTGCGATACTTCAACGCCTGGGTGCAGCCCTGCCTCGCCAGCCGCAGGTCGTCCGATTGCGAGGCGAACTCAATAAGTACGACGTGCAACACGCTCAGCCCTAAGGACGCGTCCGTCGAGCTCAAAAGTACCGCCAGCGACAACAGCTCCTTCGTCGTGTTCGCGCGCTCCGAGGGCGAAGCCCCACCGGACGGCGCCACCGGGGCCGCGCCTGAGCGTAACGACTCCACCGACAAGACGAGAGCCTCGTCCAAACGCGAGTCGAAGGACGACTTGGCTTCGTCGATGCGGAAGTCGCAAGGTGACGTGACGTCATCGACAGAGCAGACGGACTGCGGGCTGCGGCTGGAGCTCTACATACAGATGCAGCTCTGCGACCCGAACAGCCTCTACGACTGGCTGCGGAACAACCGCGACTGCCCCGACCGATACCAGCGGGCGAAAGTGATATTCGGTCAGATCGTGTCTGCGGTCGAGTACGTCCACCTTGAGGGGCTGATCCACCGCGACCTCAAGCCCAGCAACATAATGTTCGCGCCCAACGGCCAGGTCAAGGTGGCGGACTTCGGGCTCGTGGCGTACATGCACGACGCGCCCGCACTCTGCGGCGCCGCCCGCACGCCGCGCGCGCACCCGCAGCTCACGCACAGAGTCGGCACGGAAGTGTACATGTCGCCGGAGCAGCTGCAGCGGCGCCCGTACAGCTACAAGGTGGACATATACTCGCTCGGCCTCATCCTGCTCGAGCTGCTGCAGCCCTTCGGTACGGAGGCGGAGCGCGCCGCCTGCCTCACCAGGGTCCGGGACCAAGTCTACCCGGACAGCTTCGAAGATAAATACCCTAAAGAGTGA
Protein
MLDQFGLMKILSTIIALSYLVCYLQAEDVPLQLPHCNLSKTKGSSLNNYVMVSTLDGRLTAFNLENGTKAWVVETPPLLSSNLASSEFSGENRVKLVPSLRGSLYTVHGSKIKRLPFNIHQLLSESFTESNNLITAGGHDILRIGIDTKTGRIIYKCNSNGCNNEQQTTETGGDVLVLTRQATAIRGLDPRTGIEKWNFSTAAHELQISRGECIGDRTVTESPVNMHVSMHNQLVTVLSGDLNKPLWEHKLDAPITDVWELYDGNLRNSLWQDSRSSNIMGQMLYVGDHQEQLYILDEFDHSQKDRSLMVVPYQSRKRFIILDKENVYLPLSDKSTYIPFHEAEDSNATEVSDKVKKVRSWLFDILTCFTLALALVLLIFPFYARIRVMITNVPQTTVNVVFNEDIPSEKELEAKRPSNPEYPGRYEKDFTTVRRLGRGGFGVVFEARNNIDNCSYAVKRITLPRKKERHERVLREARALANLEHPHIVRYFNAWVQPCLASRRSSDCEANSISTTCNTLSPKDASVELKSTASDNSSFVVFARSEGEAPPDGATGAAPERNDSTDKTRASSKRESKDDLASSMRKSQGDVTSSTEQTDCGLRLELYIQMQLCDPNSLYDWLRNNRDCPDRYQRAKVIFGQIVSAVEYVHLEGLIHRDLKPSNIMFAPNGQVKVADFGLVAYMHDAPALCGAARTPRAHPQLTHRVGTEVYMSPEQLQRRPYSYKVDIYSLGLILLELLQPFGTEAERAACLTRVRDQVYPDSFEDKYPKE
Summary
Uniprot
EMBL
BABH01039427
BABH01039428
BABH01039429
BABH01039430
NWSH01000139
PCG79151.1
+ More
KQ459995 KPJ18535.1 KQ459600 KPI94292.1 GFDF01004872 JAV09212.1 GFDF01004871 JAV09213.1 GEZM01087803 JAV58468.1 GBYB01001331 GBYB01001333 JAG71098.1 JAG71100.1 JXLN01001862 KPM02416.1 HACA01028785 CDW46146.1 CAEY01000675 GEZM01087802 JAV58470.1 MTYJ01000004 OQV24909.1 GECU01004820 JAT02887.1 CM000364 EDX11719.1
KQ459995 KPJ18535.1 KQ459600 KPI94292.1 GFDF01004872 JAV09212.1 GFDF01004871 JAV09213.1 GEZM01087803 JAV58468.1 GBYB01001331 GBYB01001333 JAG71098.1 JAG71100.1 JXLN01001862 KPM02416.1 HACA01028785 CDW46146.1 CAEY01000675 GEZM01087802 JAV58470.1 MTYJ01000004 OQV24909.1 GECU01004820 JAT02887.1 CM000364 EDX11719.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
3QD2
E-value=6.77197e-43,
Score=441
Ontologies
PATHWAY
GO
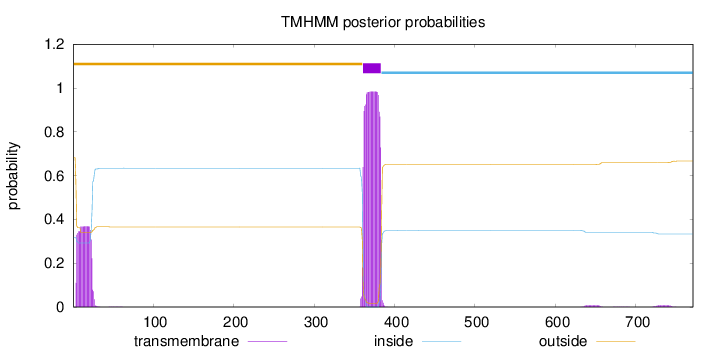
Topology
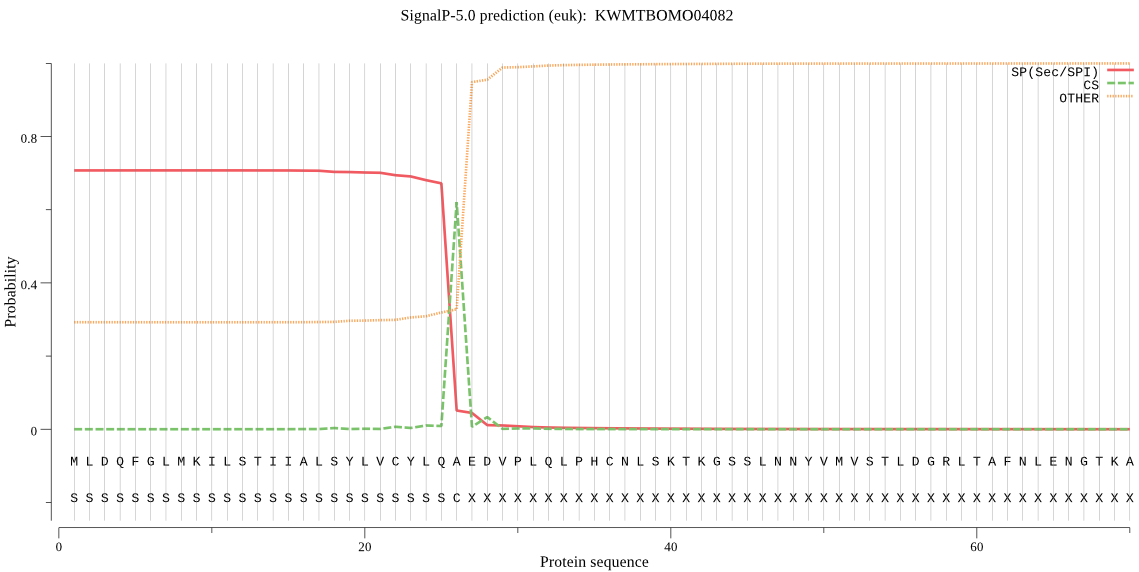
SignalP
Position: 1 - 26,
Likelihood: 0.708316
Length:
771
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
29.83525
Exp number, first 60 AAs:
7.31621
Total prob of N-in:
0.31860
outside
1 - 360
TMhelix
361 - 383
inside
384 - 771
Population Genetic Test Statistics
Pi
233.254328
Theta
180.71094
Tajima's D
0.974951
CLR
0.17415
CSRT
0.653817309134543
Interpretation
Uncertain
