Gene
KWMTBOMO04076 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008621
Annotation
3-hydroxyacyl-CoA_dehydrogenase?_partial_[Agrotis_segetum]
Full name
3-hydroxyacyl-CoA dehydrogenase type-2
Alternative Name
17-beta-hydroxysteroid dehydrogenase 10
3-hydroxy-2-methylbutyryl-CoA dehydrogenase
3-hydroxyacyl-CoA dehydrogenase type II
Mitochondrial ribonuclease P protein 2
Scully protein
Type II HADH
3-hydroxy-2-methylbutyryl-CoA dehydrogenase
3-hydroxyacyl-CoA dehydrogenase type II
Mitochondrial ribonuclease P protein 2
Scully protein
Type II HADH
Location in the cell
Mitochondrial Reliability : 1.828
Sequence
CDS
ATGCATAAGGGTATGGTCAGCCTGGTAACTGGTGGAGCTTCTGGGTTAGGTAGGGCTACAGCTGAGAGACTGGTGAAACAAGGCGGAAAGGTTGTTCTGCTAGATATACAAGGAACGAGACTTAAAGAAGTTACAAAAGAAATAGGGGACGATGCTGTAGCTGTGACGGGATCAATAACATCAGAGGATGATGTTAAAAAAGCACTGGACATAACCAAAGACAAATTTGGCAGACTTGATACACTAGTCAATTGCGCTGGACAGTCAATAACACATCAGATTTATAACTTTAATAAAGATTTACCCAGTACATTGGCAGATTTTACAAAAATTATCAATATCAACACTGTTGGAACATTCAACACAATTAGGTTATCGGCTGGACTGATTGGTAAGAATGAACCAGACGCTAATGGCCAAAGAGGAGTTATAGTAAATACGGCATCAACAATAGCATACGAAGGAGATATTGGTCAAGCTGCATATGCGGCATCAACTGCGGCAATAATAGGAATGACGTTACCGATTGCAAGGGATCTAGCTTCAAAAGGCATTCGAGTTGTTACCATAGCACCAGGTATATTTGACACGCCCCTAACGTCATATTTACCAGAGAAAATGAGGGAATTCATCAAGCGTATGACTCCATTCCCCTCGAGATTGGGCAAACCTGAGGAGTTTGCTCATCTCGTTACAACTATAGTTGAGAATCCAATGTTGAACGGAGAAGTCATAAGATTAGATGGTGCACAAAGGTGGTTTCCGCTTTGA
Protein
MHKGMVSLVTGGASGLGRATAERLVKQGGKVVLLDIQGTRLKEVTKEIGDDAVAVTGSITSEDDVKKALDITKDKFGRLDTLVNCAGQSITHQIYNFNKDLPSTLADFTKIININTVGTFNTIRLSAGLIGKNEPDANGQRGVIVNTASTIAYEGDIGQAAYAASTAAIIGMTLPIARDLASKGIRVVTIAPGIFDTPLTSYLPEKMREFIKRMTPFPSRLGKPEEFAHLVTTIVENPMLNGEVIRLDGAQRWFPL
Summary
Description
May function in mitochondrial tRNA maturation. Catalyzes the beta-oxidation at position 17 of androgens and estrogens, and has 3-alpha-hydroxysteroid dehydrogenase activity with androsterone. Catalyzes the third step in the beta-oxidation of fatty acids. Carries out oxidative conversions of 7-beta-hydroxylated bile acids. Also exhibits 20-beta-OH and 21-OH dehydrogenase activities with C21 steroids. Required for cell survival during embryonic development. May play a role in germline formation.
Catalytic Activity
a (3S)-hydroxyacyl-CoA + NAD(+) = a 3-oxoacyl-CoA + H(+) + NADH
(2S,3S)-3-hydroxy-2-methylbutanoyl-CoA + NAD(+) = 2-methyl-3-oxobutanoyl-CoA + H(+) + NADH
NAD(+) + testosterone = androst-4-ene-3,17-dione + H(+) + NADH
NADP(+) + testosterone = androst-4-ene-3,17-dione + H(+) + NADPH
(2S,3S)-3-hydroxy-2-methylbutanoyl-CoA + NAD(+) = 2-methyl-3-oxobutanoyl-CoA + H(+) + NADH
NAD(+) + testosterone = androst-4-ene-3,17-dione + H(+) + NADH
NADP(+) + testosterone = androst-4-ene-3,17-dione + H(+) + NADPH
Biophysicochemical Properties
33.7 uM for acetoacetyl-CoA (in the presence of 0.2 mM NADH, at pH 6.4 and 25 degrees Celsius)
101 uM for beta-hydroxybutyryl-CoA (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
37.3 uM for androsterone (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
12.3 uM for 5-alpha-dihydrotestosterone (in the presence of 0.2 mM NADH, at pH 6.4 and 25 degrees Celsius)
11.1 uM for 17-beta-estradiol (in the presence of 0.2 mM NADH, at pH 9.3 and 25 degrees Celsius)
9 uM for 5-alpha-pregnan-20-beta-ol-3-one (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
3 uM for isoursodeoxycholic acid (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
32.5 uM for NADH (in the presence of acetoacetyl-CoA, at pH 7.0 and 25 degrees Celsius)
64.4 uM for NAD (in the presence of beta-hydroxybutyryl-CoA, at pH 9.3 and 25 degrees Celsius)
124 uM for NAD (in the presence of aldosterone, at pH 9.3 and 25 degrees Celsius)
101 uM for beta-hydroxybutyryl-CoA (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
37.3 uM for androsterone (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
12.3 uM for 5-alpha-dihydrotestosterone (in the presence of 0.2 mM NADH, at pH 6.4 and 25 degrees Celsius)
11.1 uM for 17-beta-estradiol (in the presence of 0.2 mM NADH, at pH 9.3 and 25 degrees Celsius)
9 uM for 5-alpha-pregnan-20-beta-ol-3-one (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
3 uM for isoursodeoxycholic acid (in the presence of 1 mM NAD, at pH 9.3 and 25 degrees Celsius)
32.5 uM for NADH (in the presence of acetoacetyl-CoA, at pH 7.0 and 25 degrees Celsius)
64.4 uM for NAD (in the presence of beta-hydroxybutyryl-CoA, at pH 9.3 and 25 degrees Celsius)
124 uM for NAD (in the presence of aldosterone, at pH 9.3 and 25 degrees Celsius)
Subunit
Multimer.
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
Complete proteome
Mitochondrion
NAD
Oxidoreductase
Reference proteome
tRNA processing
Feature
chain 3-hydroxyacyl-CoA dehydrogenase type-2
Uniprot
H9JGH4
A0A068FMP4
A0A194PLU3
A0A3S2NNC5
A0A194RMH3
A0A212EJ30
+ More
A0A2A4K558 B4Q2N6 B3NTA4 A0A1E1XB23 B4M6R1 A0A1W4UN75 O18404 R4V243 B4I6N3 A0A224Z8S0 A0A2C9JID1 L7MDN0 A0A0M4EZZ4 A0A3B0K0K0 A0A293MXH6 A0A0C9RY70 B3MXV0 B4L296 Q29H71 B4H078 D6WFE2 A0A1B0GJR4 A0A1W4XN13 G3MK41 A0A131YYH0 A0A1D2M829 B4JNB1 A0A0A1X8C9 A0A1L8DZJ3 B4NEB5 E3TDR7 A0A023FVV6 A0A131XC52 E3TDJ4 A0A2R5LN65 A0A069DY28 A0A0P4VTY5 R4FNB7 A0A034WG91 A0A0C9R451 U3GM45 T1PE05 A0A0K8UAH4 A0A0A9Z6D1 Q5XJS8 A0A067R1F2 A0A3B4C668 A0A1Z5L9R7 A0A1W4XXI0 W8C108 A0A023FLX5 A0A1A9XA65 A0A0K8RNJ1 M7AQR3 A0A1W4XJI9 A0A1I8P3Q2 A0A2D0SH21 A0A1B6LJU8 A0A1B6F172 A0A224XV14 A0A3B3C5E4 A0A1I8P3Q5 D3TLT8 I3KID3 A0A3S0ZQZ8 A0A1B6JZI7 A0A3B4H904 A0A023F9Q1 A0A0V0G444 A0A3P9DNA6 A0A3B4WFP0 A0A3B4TR48 A0A3B4WH98 A0A3B4TR38 A0A1A7XM58 U5EUU3 H3B6K3 A0A3Q3L724 A0A0L8HI08 A0A3Q2DZR4 A0A3B1KFP3 A0A3Q2Y663 A0A2M3ZCD3 A0A2M4BXJ7 A0A3Q2VAE6 A0A3P8PZC7 A0A182FA62 A0A3Q2VJS8 A0A3B3HLN0 A0A3P9IJ16 A0A0F8ASV8 A0A1L8EE35 A0A2M3ZCB0 A0A1Q3G3T0
A0A2A4K558 B4Q2N6 B3NTA4 A0A1E1XB23 B4M6R1 A0A1W4UN75 O18404 R4V243 B4I6N3 A0A224Z8S0 A0A2C9JID1 L7MDN0 A0A0M4EZZ4 A0A3B0K0K0 A0A293MXH6 A0A0C9RY70 B3MXV0 B4L296 Q29H71 B4H078 D6WFE2 A0A1B0GJR4 A0A1W4XN13 G3MK41 A0A131YYH0 A0A1D2M829 B4JNB1 A0A0A1X8C9 A0A1L8DZJ3 B4NEB5 E3TDR7 A0A023FVV6 A0A131XC52 E3TDJ4 A0A2R5LN65 A0A069DY28 A0A0P4VTY5 R4FNB7 A0A034WG91 A0A0C9R451 U3GM45 T1PE05 A0A0K8UAH4 A0A0A9Z6D1 Q5XJS8 A0A067R1F2 A0A3B4C668 A0A1Z5L9R7 A0A1W4XXI0 W8C108 A0A023FLX5 A0A1A9XA65 A0A0K8RNJ1 M7AQR3 A0A1W4XJI9 A0A1I8P3Q2 A0A2D0SH21 A0A1B6LJU8 A0A1B6F172 A0A224XV14 A0A3B3C5E4 A0A1I8P3Q5 D3TLT8 I3KID3 A0A3S0ZQZ8 A0A1B6JZI7 A0A3B4H904 A0A023F9Q1 A0A0V0G444 A0A3P9DNA6 A0A3B4WFP0 A0A3B4TR48 A0A3B4WH98 A0A3B4TR38 A0A1A7XM58 U5EUU3 H3B6K3 A0A3Q3L724 A0A0L8HI08 A0A3Q2DZR4 A0A3B1KFP3 A0A3Q2Y663 A0A2M3ZCD3 A0A2M4BXJ7 A0A3Q2VAE6 A0A3P8PZC7 A0A182FA62 A0A3Q2VJS8 A0A3B3HLN0 A0A3P9IJ16 A0A0F8ASV8 A0A1L8EE35 A0A2M3ZCB0 A0A1Q3G3T0
EC Number
1.1.1.35
Pubmed
19121390
26385554
26354079
22118469
17994087
17550304
+ More
28503490 9585418 10731132 12537572 12537569 12917011 16979555 17924685 28797301 15562597 25576852 26131772 15632085 18362917 19820115 22216098 26830274 27289101 25830018 20634964 28049606 26334808 27129103 25348373 25315136 25401762 26823975 23594743 24845553 28528879 24495485 23624526 29451363 20353571 25186727 25474469 9215903 25329095 17554307 25835551
28503490 9585418 10731132 12537572 12537569 12917011 16979555 17924685 28797301 15562597 25576852 26131772 15632085 18362917 19820115 22216098 26830274 27289101 25830018 20634964 28049606 26334808 27129103 25348373 25315136 25401762 26823975 23594743 24845553 28528879 24495485 23624526 29451363 20353571 25186727 25474469 9215903 25329095 17554307 25835551
EMBL
BABH01039421
KJ622101
AID66691.1
KQ459600
KPI94297.1
RSAL01000210
+ More
RVE44222.1 KQ459995 KPJ18530.1 AGBW02014538 OWR41485.1 NWSH01000139 PCG79156.1 CM000162 EDX02677.1 CH954180 EDV46555.1 GFAC01002754 JAT96434.1 CH940653 EDW62478.1 Y15102 AE014298 AY121672 BT029045 BT132763 KC740963 AGM32787.1 CH480823 EDW55981.1 GFPF01011746 MAA22892.1 GACK01002974 JAA62060.1 CP012528 ALC49867.1 OUUW01000003 SPP78541.1 GFWV01019322 MAA44050.1 GBZX01000104 JAG92636.1 CH902630 EDV38565.1 CH933810 EDW07757.1 CH379064 EAL31887.1 CH479200 EDW29673.1 KQ971319 EFA00491.1 AJWK01022146 JO842242 AEO33859.1 GEDV01004318 JAP84239.1 LJIJ01002921 ODM89092.1 CH916371 EDV92204.1 GBXI01007369 JAD06923.1 GFDF01002382 JAV11702.1 CH964239 EDW82084.1 GU588497 ADO28453.1 GBBL01001701 JAC25619.1 GEFH01004639 JAP63942.1 GU588424 ADO28380.1 GGLE01006818 MBY10944.1 GBGD01002615 JAC86274.1 GDKW01000596 JAI55999.1 ACPB03014675 GAHY01000628 JAA76882.1 GAKP01005605 JAC53347.1 GBYB01001591 JAG71358.1 KF031120 AGT97387.1 KA646375 AFP61004.1 GDHF01028999 JAI23315.1 GBHO01003590 GBRD01011971 GBRD01011970 GDHC01009188 JAG40014.1 JAG53853.1 JAQ09441.1 CR735130 BC083219 BC165246 AAH83219.1 AAI65246.1 KK852996 KDR12703.1 GFJQ02002841 JAW04129.1 GAMC01003782 JAC02774.1 GBBK01001865 JAC22617.1 GADI01001659 JAA72149.1 KB575110 EMP26949.1 GEBQ01016088 JAT23889.1 GECZ01025777 JAS43992.1 GFTR01004176 JAW12250.1 CCAG010012474 EZ422390 ADD18666.1 AERX01043880 RQTK01000374 RUS80743.1 GECU01003373 JAT04334.1 GBBI01001059 JAC17653.1 GECL01003313 JAP02811.1 HADW01017781 HADX01009706 SBP19181.1 GANO01001305 JAB58566.1 AFYH01083165 AFYH01083166 AFYH01083167 AFYH01083168 AFYH01083169 AFYH01083170 AFYH01083171 KQ418094 KOF88861.1 GGFM01005367 MBW26118.1 GGFJ01008583 MBW57724.1 KQ040919 KKF33011.1 GFDG01001832 JAV16967.1 GGFM01005390 MBW26141.1 GFDL01000582 JAV34463.1
RVE44222.1 KQ459995 KPJ18530.1 AGBW02014538 OWR41485.1 NWSH01000139 PCG79156.1 CM000162 EDX02677.1 CH954180 EDV46555.1 GFAC01002754 JAT96434.1 CH940653 EDW62478.1 Y15102 AE014298 AY121672 BT029045 BT132763 KC740963 AGM32787.1 CH480823 EDW55981.1 GFPF01011746 MAA22892.1 GACK01002974 JAA62060.1 CP012528 ALC49867.1 OUUW01000003 SPP78541.1 GFWV01019322 MAA44050.1 GBZX01000104 JAG92636.1 CH902630 EDV38565.1 CH933810 EDW07757.1 CH379064 EAL31887.1 CH479200 EDW29673.1 KQ971319 EFA00491.1 AJWK01022146 JO842242 AEO33859.1 GEDV01004318 JAP84239.1 LJIJ01002921 ODM89092.1 CH916371 EDV92204.1 GBXI01007369 JAD06923.1 GFDF01002382 JAV11702.1 CH964239 EDW82084.1 GU588497 ADO28453.1 GBBL01001701 JAC25619.1 GEFH01004639 JAP63942.1 GU588424 ADO28380.1 GGLE01006818 MBY10944.1 GBGD01002615 JAC86274.1 GDKW01000596 JAI55999.1 ACPB03014675 GAHY01000628 JAA76882.1 GAKP01005605 JAC53347.1 GBYB01001591 JAG71358.1 KF031120 AGT97387.1 KA646375 AFP61004.1 GDHF01028999 JAI23315.1 GBHO01003590 GBRD01011971 GBRD01011970 GDHC01009188 JAG40014.1 JAG53853.1 JAQ09441.1 CR735130 BC083219 BC165246 AAH83219.1 AAI65246.1 KK852996 KDR12703.1 GFJQ02002841 JAW04129.1 GAMC01003782 JAC02774.1 GBBK01001865 JAC22617.1 GADI01001659 JAA72149.1 KB575110 EMP26949.1 GEBQ01016088 JAT23889.1 GECZ01025777 JAS43992.1 GFTR01004176 JAW12250.1 CCAG010012474 EZ422390 ADD18666.1 AERX01043880 RQTK01000374 RUS80743.1 GECU01003373 JAT04334.1 GBBI01001059 JAC17653.1 GECL01003313 JAP02811.1 HADW01017781 HADX01009706 SBP19181.1 GANO01001305 JAB58566.1 AFYH01083165 AFYH01083166 AFYH01083167 AFYH01083168 AFYH01083169 AFYH01083170 AFYH01083171 KQ418094 KOF88861.1 GGFM01005367 MBW26118.1 GGFJ01008583 MBW57724.1 KQ040919 KKF33011.1 GFDG01001832 JAV16967.1 GGFM01005390 MBW26141.1 GFDL01000582 JAV34463.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000007151
UP000218220
+ More
UP000002282 UP000008711 UP000008792 UP000192221 UP000000803 UP000001292 UP000076420 UP000092553 UP000268350 UP000007801 UP000009192 UP000001819 UP000008744 UP000007266 UP000092461 UP000192224 UP000094527 UP000001070 UP000007798 UP000221080 UP000015103 UP000095301 UP000000437 UP000027135 UP000261440 UP000092443 UP000031443 UP000192223 UP000095300 UP000261560 UP000092444 UP000005207 UP000271974 UP000261460 UP000265160 UP000261360 UP000261420 UP000008672 UP000261640 UP000053454 UP000265020 UP000018467 UP000264820 UP000264840 UP000265100 UP000069272 UP000001038 UP000265180 UP000265200
UP000002282 UP000008711 UP000008792 UP000192221 UP000000803 UP000001292 UP000076420 UP000092553 UP000268350 UP000007801 UP000009192 UP000001819 UP000008744 UP000007266 UP000092461 UP000192224 UP000094527 UP000001070 UP000007798 UP000221080 UP000015103 UP000095301 UP000000437 UP000027135 UP000261440 UP000092443 UP000031443 UP000192223 UP000095300 UP000261560 UP000092444 UP000005207 UP000271974 UP000261460 UP000265160 UP000261360 UP000261420 UP000008672 UP000261640 UP000053454 UP000265020 UP000018467 UP000264820 UP000264840 UP000265100 UP000069272 UP000001038 UP000265180 UP000265200
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JGH4
A0A068FMP4
A0A194PLU3
A0A3S2NNC5
A0A194RMH3
A0A212EJ30
+ More
A0A2A4K558 B4Q2N6 B3NTA4 A0A1E1XB23 B4M6R1 A0A1W4UN75 O18404 R4V243 B4I6N3 A0A224Z8S0 A0A2C9JID1 L7MDN0 A0A0M4EZZ4 A0A3B0K0K0 A0A293MXH6 A0A0C9RY70 B3MXV0 B4L296 Q29H71 B4H078 D6WFE2 A0A1B0GJR4 A0A1W4XN13 G3MK41 A0A131YYH0 A0A1D2M829 B4JNB1 A0A0A1X8C9 A0A1L8DZJ3 B4NEB5 E3TDR7 A0A023FVV6 A0A131XC52 E3TDJ4 A0A2R5LN65 A0A069DY28 A0A0P4VTY5 R4FNB7 A0A034WG91 A0A0C9R451 U3GM45 T1PE05 A0A0K8UAH4 A0A0A9Z6D1 Q5XJS8 A0A067R1F2 A0A3B4C668 A0A1Z5L9R7 A0A1W4XXI0 W8C108 A0A023FLX5 A0A1A9XA65 A0A0K8RNJ1 M7AQR3 A0A1W4XJI9 A0A1I8P3Q2 A0A2D0SH21 A0A1B6LJU8 A0A1B6F172 A0A224XV14 A0A3B3C5E4 A0A1I8P3Q5 D3TLT8 I3KID3 A0A3S0ZQZ8 A0A1B6JZI7 A0A3B4H904 A0A023F9Q1 A0A0V0G444 A0A3P9DNA6 A0A3B4WFP0 A0A3B4TR48 A0A3B4WH98 A0A3B4TR38 A0A1A7XM58 U5EUU3 H3B6K3 A0A3Q3L724 A0A0L8HI08 A0A3Q2DZR4 A0A3B1KFP3 A0A3Q2Y663 A0A2M3ZCD3 A0A2M4BXJ7 A0A3Q2VAE6 A0A3P8PZC7 A0A182FA62 A0A3Q2VJS8 A0A3B3HLN0 A0A3P9IJ16 A0A0F8ASV8 A0A1L8EE35 A0A2M3ZCB0 A0A1Q3G3T0
A0A2A4K558 B4Q2N6 B3NTA4 A0A1E1XB23 B4M6R1 A0A1W4UN75 O18404 R4V243 B4I6N3 A0A224Z8S0 A0A2C9JID1 L7MDN0 A0A0M4EZZ4 A0A3B0K0K0 A0A293MXH6 A0A0C9RY70 B3MXV0 B4L296 Q29H71 B4H078 D6WFE2 A0A1B0GJR4 A0A1W4XN13 G3MK41 A0A131YYH0 A0A1D2M829 B4JNB1 A0A0A1X8C9 A0A1L8DZJ3 B4NEB5 E3TDR7 A0A023FVV6 A0A131XC52 E3TDJ4 A0A2R5LN65 A0A069DY28 A0A0P4VTY5 R4FNB7 A0A034WG91 A0A0C9R451 U3GM45 T1PE05 A0A0K8UAH4 A0A0A9Z6D1 Q5XJS8 A0A067R1F2 A0A3B4C668 A0A1Z5L9R7 A0A1W4XXI0 W8C108 A0A023FLX5 A0A1A9XA65 A0A0K8RNJ1 M7AQR3 A0A1W4XJI9 A0A1I8P3Q2 A0A2D0SH21 A0A1B6LJU8 A0A1B6F172 A0A224XV14 A0A3B3C5E4 A0A1I8P3Q5 D3TLT8 I3KID3 A0A3S0ZQZ8 A0A1B6JZI7 A0A3B4H904 A0A023F9Q1 A0A0V0G444 A0A3P9DNA6 A0A3B4WFP0 A0A3B4TR48 A0A3B4WH98 A0A3B4TR38 A0A1A7XM58 U5EUU3 H3B6K3 A0A3Q3L724 A0A0L8HI08 A0A3Q2DZR4 A0A3B1KFP3 A0A3Q2Y663 A0A2M3ZCD3 A0A2M4BXJ7 A0A3Q2VAE6 A0A3P8PZC7 A0A182FA62 A0A3Q2VJS8 A0A3B3HLN0 A0A3P9IJ16 A0A0F8ASV8 A0A1L8EE35 A0A2M3ZCB0 A0A1Q3G3T0
PDB
1U7T
E-value=1.7489e-76,
Score=725
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion
Localizes to the lipid droplet fraction in early embryos. With evidence from 4 publications.
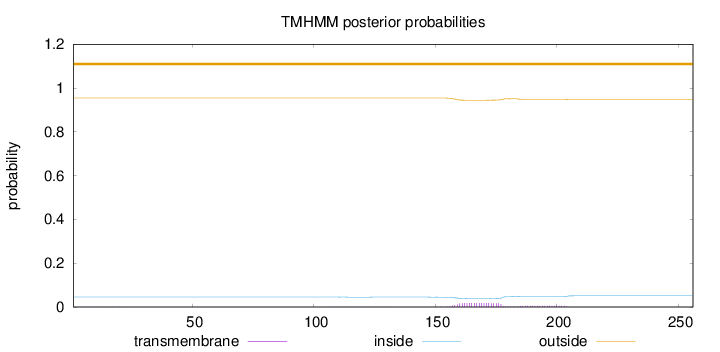
Length:
256
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.48421
Exp number, first 60 AAs:
0.0064
Total prob of N-in:
0.04553
outside
1 - 256
Population Genetic Test Statistics
Pi
282.60588
Theta
159.543585
Tajima's D
1.832725
CLR
0.000001
CSRT
0.854357282135893
Interpretation
Uncertain
