Gene
KWMTBOMO04072
Annotation
hypothetical_protein_KGM_11869_[Danaus_plexippus]
Full name
Guanylate cyclase
Location in the cell
Extracellular Reliability : 3.618
Sequence
CDS
ATGTGGTTATCTGTGTTGCTAGTGGTGTGCGCGATCCCTTGGGGTGAATCGCAGAACTGCCGCGGGGAGAAACCTCGGCTCCGCGATTGCGATCGTCTCTGCGACAAGCAGGGCAACTGTAAGATCAAAGCTGCCCTACTTCTCCCGAAGAACATAACGTTCGGAGCGTGTATCGGTGCTGTGGAGCCAGTGTTGGAGCTGGCTATGCAGCATCCGGCGGTCCAGGATGCCTTTCCCTCGTGGGTCGGCTTCGAGTGGCAGACTTATGACGTCACAGATTGCGATGCAGCATATGCTGTCATTTCCGCCATCGACGCGTATAACGACTGCGCCCATGTCTTCCTCGGTCCCGCCTGTGATTTCGCATTAGGTGATTCTATTTGA
Protein
MWLSVLLVVCAIPWGESQNCRGEKPRLRDCDRLCDKQGNCKIKAALLLPKNITFGACIGAVEPVLELAMQHPAVQDAFPSWVGFEWQTYDVTDCDAAYAVISAIDAYNDCAHVFLGPACDFALGDSI
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Feature
chain Guanylate cyclase
Uniprot
A0A2A4IZR6
A0A212FC87
A0A1E1WT37
A0A0L7LK13
A0A194PT40
A0A194RL17
+ More
B4NBA5 A0A0C9QYW7 A0A2A3EEE4 A0A0A1X5Z0 A0A088AFH3 A0A151WKS9 B4LX88 A0A0C9PWJ7 A0A1W4VFG5 A0A195EDG4 A0A195F3D4 A0A0P9CAM4 B3P3W8 A0A310SF44 A0A158NDF8 A0A195B4Z8 Q9VF16 E9J769 F4W973 A0A0K8V105 W5JDF4 B4PRJ1 A0A232FFJ9 A0A151IFB5 A0A2M4CHA1 B4QXT3 B4HLG8 A0A0Q9XA14 K7J4N3 B4GEV3 Q296P8 A0A0L7QKR9 A0A026WAX4 A0A3B0KKH2 A0A154PID3 B4JT48 A0A1A9WJI8 E2B8H8 A0A182FSG2 A0A1B0BBW1 A0A1A9YK42 A0A1A9ZTC9 A0A1S4G740 E0VPJ2 A0A1B0G6S2 A0A182T9U6 A0A1B6MMP7 A0A182JED6 A0A182MJZ5 A0A1W4WJN3 A0A1W4W9J9 A0A1Y1M0M0 A0A336LM52 A0A0L8G872 D6W7W3
B4NBA5 A0A0C9QYW7 A0A2A3EEE4 A0A0A1X5Z0 A0A088AFH3 A0A151WKS9 B4LX88 A0A0C9PWJ7 A0A1W4VFG5 A0A195EDG4 A0A195F3D4 A0A0P9CAM4 B3P3W8 A0A310SF44 A0A158NDF8 A0A195B4Z8 Q9VF16 E9J769 F4W973 A0A0K8V105 W5JDF4 B4PRJ1 A0A232FFJ9 A0A151IFB5 A0A2M4CHA1 B4QXT3 B4HLG8 A0A0Q9XA14 K7J4N3 B4GEV3 Q296P8 A0A0L7QKR9 A0A026WAX4 A0A3B0KKH2 A0A154PID3 B4JT48 A0A1A9WJI8 E2B8H8 A0A182FSG2 A0A1B0BBW1 A0A1A9YK42 A0A1A9ZTC9 A0A1S4G740 E0VPJ2 A0A1B0G6S2 A0A182T9U6 A0A1B6MMP7 A0A182JED6 A0A182MJZ5 A0A1W4WJN3 A0A1W4W9J9 A0A1Y1M0M0 A0A336LM52 A0A0L8G872 D6W7W3
EC Number
4.6.1.2
Pubmed
EMBL
NWSH01004234
PCG65315.1
AGBW02009216
OWR51327.1
GDQN01000859
GDQN01000188
+ More
JAT90195.1 JAT90866.1 JTDY01000846 KOB75684.1 KQ459600 KPI94300.1 KQ459995 KPJ18528.1 CH964232 EDW81069.2 GBYB01005872 JAG75639.1 KZ288287 PBC29381.1 GBXI01008204 JAD06088.1 KQ983002 KYQ48417.1 CH940650 EDW66740.2 GBYB01005873 JAG75640.1 KQ979039 KYN23265.1 KQ981855 KYN34896.1 CH902617 KPU80309.1 CH954181 EDV49074.1 KQ777939 OAD52020.1 ADTU01001012 KQ976598 KYM79571.1 AE014297 AAF55245.1 GL768421 EFZ11328.1 GL888002 EGI69255.1 GDHF01019720 JAI32594.1 ADMH02001856 ETN60844.1 CM000160 EDW97391.2 NNAY01000279 OXU29534.1 KQ977800 KYM99574.1 GGFL01000353 MBW64531.1 CM000364 EDX12757.1 CH480815 EDW41988.1 CH933806 KRG01393.1 AAZX01000467 CH479182 EDW34138.1 CM000070 EAL28409.3 KQ414940 KOC59165.1 KK107293 EZA53252.1 OUUW01000008 SPP84278.1 KQ434902 KZC11078.1 CH916373 EDV94938.1 GL446332 EFN88012.1 JXJN01011716 DS235366 EEB15298.1 CCAG010013864 GEBQ01002838 JAT37139.1 AXCM01003272 GEZM01047698 JAV76777.1 UFQT01000054 SSX19066.1 KQ423235 KOF73226.1 KQ971307 EFA11145.1
JAT90195.1 JAT90866.1 JTDY01000846 KOB75684.1 KQ459600 KPI94300.1 KQ459995 KPJ18528.1 CH964232 EDW81069.2 GBYB01005872 JAG75639.1 KZ288287 PBC29381.1 GBXI01008204 JAD06088.1 KQ983002 KYQ48417.1 CH940650 EDW66740.2 GBYB01005873 JAG75640.1 KQ979039 KYN23265.1 KQ981855 KYN34896.1 CH902617 KPU80309.1 CH954181 EDV49074.1 KQ777939 OAD52020.1 ADTU01001012 KQ976598 KYM79571.1 AE014297 AAF55245.1 GL768421 EFZ11328.1 GL888002 EGI69255.1 GDHF01019720 JAI32594.1 ADMH02001856 ETN60844.1 CM000160 EDW97391.2 NNAY01000279 OXU29534.1 KQ977800 KYM99574.1 GGFL01000353 MBW64531.1 CM000364 EDX12757.1 CH480815 EDW41988.1 CH933806 KRG01393.1 AAZX01000467 CH479182 EDW34138.1 CM000070 EAL28409.3 KQ414940 KOC59165.1 KK107293 EZA53252.1 OUUW01000008 SPP84278.1 KQ434902 KZC11078.1 CH916373 EDV94938.1 GL446332 EFN88012.1 JXJN01011716 DS235366 EEB15298.1 CCAG010013864 GEBQ01002838 JAT37139.1 AXCM01003272 GEZM01047698 JAV76777.1 UFQT01000054 SSX19066.1 KQ423235 KOF73226.1 KQ971307 EFA11145.1
Proteomes
UP000218220
UP000007151
UP000037510
UP000053268
UP000053240
UP000007798
+ More
UP000242457 UP000005203 UP000075809 UP000008792 UP000192221 UP000078492 UP000078541 UP000007801 UP000008711 UP000005205 UP000078540 UP000000803 UP000007755 UP000000673 UP000002282 UP000215335 UP000078542 UP000000304 UP000001292 UP000009192 UP000002358 UP000008744 UP000001819 UP000053825 UP000053097 UP000268350 UP000076502 UP000001070 UP000091820 UP000008237 UP000069272 UP000092460 UP000092443 UP000092445 UP000009046 UP000092444 UP000075901 UP000075880 UP000075883 UP000192223 UP000053454 UP000007266
UP000242457 UP000005203 UP000075809 UP000008792 UP000192221 UP000078492 UP000078541 UP000007801 UP000008711 UP000005205 UP000078540 UP000000803 UP000007755 UP000000673 UP000002282 UP000215335 UP000078542 UP000000304 UP000001292 UP000009192 UP000002358 UP000008744 UP000001819 UP000053825 UP000053097 UP000268350 UP000076502 UP000001070 UP000091820 UP000008237 UP000069272 UP000092460 UP000092443 UP000092445 UP000009046 UP000092444 UP000075901 UP000075880 UP000075883 UP000192223 UP000053454 UP000007266
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IZR6
A0A212FC87
A0A1E1WT37
A0A0L7LK13
A0A194PT40
A0A194RL17
+ More
B4NBA5 A0A0C9QYW7 A0A2A3EEE4 A0A0A1X5Z0 A0A088AFH3 A0A151WKS9 B4LX88 A0A0C9PWJ7 A0A1W4VFG5 A0A195EDG4 A0A195F3D4 A0A0P9CAM4 B3P3W8 A0A310SF44 A0A158NDF8 A0A195B4Z8 Q9VF16 E9J769 F4W973 A0A0K8V105 W5JDF4 B4PRJ1 A0A232FFJ9 A0A151IFB5 A0A2M4CHA1 B4QXT3 B4HLG8 A0A0Q9XA14 K7J4N3 B4GEV3 Q296P8 A0A0L7QKR9 A0A026WAX4 A0A3B0KKH2 A0A154PID3 B4JT48 A0A1A9WJI8 E2B8H8 A0A182FSG2 A0A1B0BBW1 A0A1A9YK42 A0A1A9ZTC9 A0A1S4G740 E0VPJ2 A0A1B0G6S2 A0A182T9U6 A0A1B6MMP7 A0A182JED6 A0A182MJZ5 A0A1W4WJN3 A0A1W4W9J9 A0A1Y1M0M0 A0A336LM52 A0A0L8G872 D6W7W3
B4NBA5 A0A0C9QYW7 A0A2A3EEE4 A0A0A1X5Z0 A0A088AFH3 A0A151WKS9 B4LX88 A0A0C9PWJ7 A0A1W4VFG5 A0A195EDG4 A0A195F3D4 A0A0P9CAM4 B3P3W8 A0A310SF44 A0A158NDF8 A0A195B4Z8 Q9VF16 E9J769 F4W973 A0A0K8V105 W5JDF4 B4PRJ1 A0A232FFJ9 A0A151IFB5 A0A2M4CHA1 B4QXT3 B4HLG8 A0A0Q9XA14 K7J4N3 B4GEV3 Q296P8 A0A0L7QKR9 A0A026WAX4 A0A3B0KKH2 A0A154PID3 B4JT48 A0A1A9WJI8 E2B8H8 A0A182FSG2 A0A1B0BBW1 A0A1A9YK42 A0A1A9ZTC9 A0A1S4G740 E0VPJ2 A0A1B0G6S2 A0A182T9U6 A0A1B6MMP7 A0A182JED6 A0A182MJZ5 A0A1W4WJN3 A0A1W4W9J9 A0A1Y1M0M0 A0A336LM52 A0A0L8G872 D6W7W3
Ontologies
KEGG
GO
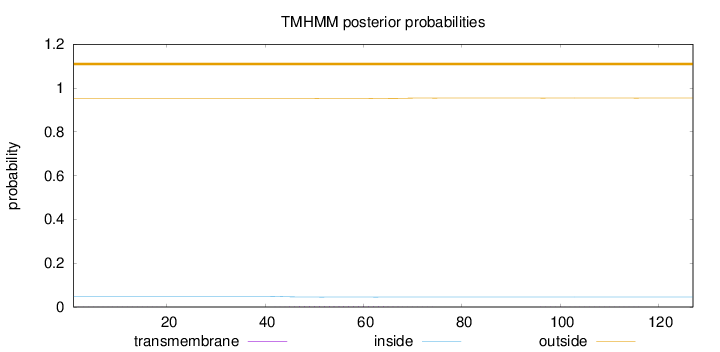
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.997326
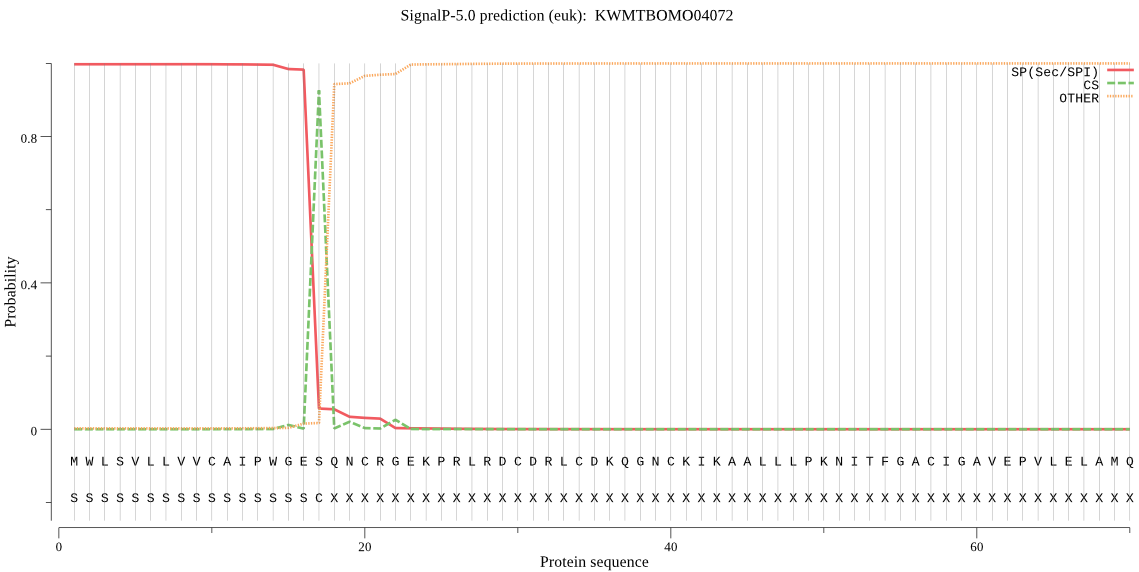
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0750399999999999
Exp number, first 60 AAs:
0.05099
Total prob of N-in:
0.04800
outside
1 - 127
Population Genetic Test Statistics
Pi
313.171678
Theta
175.025986
Tajima's D
2.775245
CLR
0.143218
CSRT
0.965551722413879
Interpretation
Uncertain
