Gene
KWMTBOMO04068
Pre Gene Modal
BGIBMGA010092
Annotation
receptor_type_guanylyl_cyclase_[Bombyx_mori]
Full name
Guanylate cyclase
Location in the cell
Nuclear Reliability : 1.558 PlasmaMembrane Reliability : 2.092
Sequence
CDS
ATGTTAGTGGAGATCGTGCAAGCGATTCTCCCCCTTATCGTTAAGAGACTAAAGTGCAAAAAACGTTCATACGGATATCGAGCGGATTACAGGAAGAGACGAAGCGAATACGCTACCGATACTCATGTTGTTTGTGACCATGTTCAAACACAGAAGATTAAAACGAGATCTGTAATTAATCATGAAACACCAATATTATTCACATTCCTGGTTCTTTACGTGTGTATTTTGAATGTCATTATTAAATGTGACGTTAGTGTTAATGATGTAAATAATAAAGACGGTTTTATTAAACATGGATACGATACGCACGTGAATTTGAGGAACAAGTCTAAATGGTTAGGGCATAGGCACGAAGATAGAGTGTACGGAAAGAATATGCGCAAAGGACATGTGTCGATGAAAAGACACACCGGTGAGAAGAAAGACGATAATTTTAGTCCAGACGAACAAGACTTCGATAAGTTTAACGATTTTTATAAGAGCAACGAAGATTTGACCAACGTTACTGAGTACCCGGATATATTTGAGATGTCGCGATACGTTTACGATGATACCCGCGCCATTGGCCATCCCCATGACTTGGCCAAAAATCCTTATTACGTGAAGAATATTAATTTTAATTTACGCCCAGAGAGGCACCAGTTCCACCGGAAGATCGTGAAACTGGGCGTTCTGTTGCCCGCCGACCCAAATCAAGTGTTTTCGTTGGTCAAGGTTCTGCCAATATTGGAAATGGCCATACCGGCTGTTACAAAGCAGGACGGACCCCTTCCTGGTTGGAAAATACTGGTGGACTACAGAGATACCCTGTGTTCTTCTGTGGAAGGACCACTGGCGGCCTTCGAGTTTTACGTTAATGGATCAGCTGATGCTTTTATTGGTCCCGGTTGCGAGTACGTCATTGCTCCGGTGGCAAGGTATGCTGGACCTTGGGGTATACCGGTGTTGACAGCGGGCGCTCAGGCTGAAGCTTTTTTCTACAAACACCCAAGTTACGCAACTCTGACCAGAATGATGGGTAGCTACACTCAAGCAGGCGTCGCTATTCGCAATATCTTCGAGGAGTTCAACTGGCGTCGCTTAGGGATGCTCTACCACAACAACGACCCCTCGTCGGGGAAGGGAAACAGCCCGTGCTTCTTGACACTGAGCGCCGTGTTCACGGTGTTGAACAAGAAAACGAGCGGATCCGACAACATTATAACGGCCCAGTTCGACGAGACGAACACGACAAATACGAAATTCAAGGAGCTCCTTCACAAACTGTCCCTGAGCACGAGAATTGTCGTGTTATGTGCGAACCCGGCTACAGTAAGAGAAATAATGCTGGCAGCCGACGATCTCAACATGGTGTCGTCCGGAGAATACGTGTTCTTTAATATTGAACTTTTTTCTAATTTAGCATCAGCTTCATCCAAGGAACCCTGGAAATCTGAGAACGACACGGAGGAAAGAAACGAAAGAGCAAGGAGGGCCTATAGTGCTGTGCTGACCGTGACGAGTCCGGCTCCAGAGAAAAAGGAATACCTTGAGTTTTCTGATCAGGTCAAAGAGCTCGCCGCAACGAAGTACAACTACACGTTCGGGAAAGGCGAAGTTGTTTCAACTTTCGTGGCTGCCTTTTACGATGCGGTCCTTCTGTACGCTCTGGCGCTGAACGACACTCTGCAGCAGGCGACCGATCCTCGAGGGCAGCTGGACGGCGCTGCGGTAATGAGGAATATGTGGAATAGGACTTTTCAAGGTATAACCGGCGAGGTTGTAATAAACAGCAACGGTGATCGCGTCGCCTCTTATTCGTTGCTCGATATGAACCCAAATACCAGCAAATTTGAGGTGGTAGCAACATACGTGGCCGCAAATAAAACACTACAGTTCACCGAGAATAGACCTATTTACTGGGCTGGCGGCCGAACCACTCCGCCCCCTGACACACCGGAATGCGGCTTCGATGGTTCGCTTTGTCCGGATAACTCACTCCCGGTTTACGCGATTCTCAGTATTGTATTGAGTTCCGTCGTTGTGATCTTGGCGATACTATCAGTCTTCATATACAGGCATTATAAACTTGAGGCGGAAATCGCGATGATGACCTGGAGGGTTAGCTGGAGTGATGTCCTGCAGCCTGGCGGTCGACTCAGAGGCAGTCTGCATTCAATAACCAGACGCTACAGCACTGGAACCACGTTCTCCGACGAAGCGACATCATTGGCTGGAGATCGCCAAGTGTACACTCCATGTGGATTCTACAAGGGCTGCAGAGTCGCAATCAAACTAGTGGACAAACAGCGAATAGACCTCACCAGGCCGTTACTGCTGGAGCTGAAGAAAATGAAAGATCTCGAGCACGACCATTTGGCGAGGTTCTATGGGGCCTGCGTTGATCCGCCGCACTGCTGCTTGTTGACCGAATACTGTCCGAAAGGATCTTTGCAGGATATCCTTGAGAATGAATCTATCAAACTAGATTGGATGTTCAAAGTCAGTTTGATGCACGATATAGTAAAGGGCATGCATTACTTACACAATTCCGACATAAAATCGCATGGAGCACTCAAGTCGAGCAATTGTGTAGTTGATTCGAGATTTGTATTAAAAATAACCGACTTCGGAATACACGCCTTAAGAACTTCGGAAAAGGACTCGAGCGCTCACAGTTATTGGACTCGTCTCCTCTGGACAGCTCCGGAACTATTGAGAATGCCAGAGCCGCCGCCGGAGGGCACACAGAAAGGCGACGTTTATTCCTTTGCTATTGTCATGCACGAGATAGTGAATAGGCAGGGAGTATTTTGGCTCGGACCCGGCATAGAATTCTCACCTAAAGAAATCATTGAAACGATCAGGGCGAGTTGTCTACGGCCTAATACGTCTCATCACCGATCTTGCGAAGCTGATGACGCGGCAGAGCTGATGCGTCGGTGCTGGGCAGAGGATCCAACTGAAAGACCTGACTTTGGAAACCTTAAGGGTGCCATAAGGAGGCTCAACAAAACTCAAGAAAGTAGCAATATTTTAGACAACCTGCTTTCCCGTATGGAACAATATGCGAATAATTTGGAGACTCTAGTTAGCGAGAGAACTCAGGACTATTTAGAAGAAAAGAAAAAATGCGAAGAGCTACTGTACCAGTTACTGCCTAAGTCAGTGGCCTCCCAGCTAATCAACGGGCAGTCAGTGGTAGCGGAAACCTTCGAACAAGTCACAATCTACTTTTCAGATATAGTTGGTTTCACTGCTCTCTCAGCTTCCTCCACTCCTATGCAAGTGGTGGACCTCCTCAATGATCTGTATACCTGTTTTGATTCCATTGTGGAAAACTATGACGTCTATAAGGTGGAAACCATAGGTGATGCGTACATGGTGGTATCTGGTCTGCCTGAACGCAACGGGACCAGACATGCGTGCGAAGTGGCGAGGATGGCTCTCGCTTTACTGGATGCCGTCAGGAGATTCCGCATCAGGCATCGGCCTATGGACCAGCTGAGACTTCGGATCGGATTACATTCTGGTCCCTGTGTTGCTGGAGTGGTGGGCTTGAAGATGCCGCGCTACTGCTTGTTCGGTGACACAGTAAACACCGCGTCTCGTATGGAATCCAACGGAGAAGCTCTGAAGATACACGTGTCTCCGGACACGAAAGCTCTGCTGGATACATTCGGGACGTTCCATTTGGAGTGCAGAGGGGAGGTCGCCATGAAGGGTAAAGGAATTCTAGTCACATATTGGTTATTGGGTGAAAAAACGGACCCAAATGCTCCGAAGGAAAAACCTTCGCTGGTCAAAAACCAAACACTTTCAACCGAAAGCGGTTCACTTTCGCATCTCACACCATTTCGGCAGAGAATGGATCAAAACTCCAGGTCTTCAAGTAGGCGTGGGTCAGGAGTTTTCCAGCAGAGGTCTATAGACGTCACCAAGCTAGAAAAAGAAACACAGCCGTTGCTTCAAAGCCTCGCGATTCAAAGGCAGCTCTCAGACCCTACGGAACCAGTAGCTAATGGTATAGACAACGAGGAATCAGACAAACTATCAGTAACTAATTCAATATCGTTAAATGTAAACTCTCTAAGCCACAATCACCAGGGGCATCCACGGGTCAAACTGAAGGAAGCCAACTCGATCAACCACAAGTCAAAAGTTGGTAATAATAATTGGATTTCAAAACTGCCTACAGCCAGTAATTCCTTTGACAACGGAACCAAGAAAGCCAGTGAGCGTCTTAAAGATTACAGCAGTGTACCGCTTCTTTCGAAGGCCGATACACCTAATGATTCTATAGTGTAA
Protein
MLVEIVQAILPLIVKRLKCKKRSYGYRADYRKRRSEYATDTHVVCDHVQTQKIKTRSVINHETPILFTFLVLYVCILNVIIKCDVSVNDVNNKDGFIKHGYDTHVNLRNKSKWLGHRHEDRVYGKNMRKGHVSMKRHTGEKKDDNFSPDEQDFDKFNDFYKSNEDLTNVTEYPDIFEMSRYVYDDTRAIGHPHDLAKNPYYVKNINFNLRPERHQFHRKIVKLGVLLPADPNQVFSLVKVLPILEMAIPAVTKQDGPLPGWKILVDYRDTLCSSVEGPLAAFEFYVNGSADAFIGPGCEYVIAPVARYAGPWGIPVLTAGAQAEAFFYKHPSYATLTRMMGSYTQAGVAIRNIFEEFNWRRLGMLYHNNDPSSGKGNSPCFLTLSAVFTVLNKKTSGSDNIITAQFDETNTTNTKFKELLHKLSLSTRIVVLCANPATVREIMLAADDLNMVSSGEYVFFNIELFSNLASASSKEPWKSENDTEERNERARRAYSAVLTVTSPAPEKKEYLEFSDQVKELAATKYNYTFGKGEVVSTFVAAFYDAVLLYALALNDTLQQATDPRGQLDGAAVMRNMWNRTFQGITGEVVINSNGDRVASYSLLDMNPNTSKFEVVATYVAANKTLQFTENRPIYWAGGRTTPPPDTPECGFDGSLCPDNSLPVYAILSIVLSSVVVILAILSVFIYRHYKLEAEIAMMTWRVSWSDVLQPGGRLRGSLHSITRRYSTGTTFSDEATSLAGDRQVYTPCGFYKGCRVAIKLVDKQRIDLTRPLLLELKKMKDLEHDHLARFYGACVDPPHCCLLTEYCPKGSLQDILENESIKLDWMFKVSLMHDIVKGMHYLHNSDIKSHGALKSSNCVVDSRFVLKITDFGIHALRTSEKDSSAHSYWTRLLWTAPELLRMPEPPPEGTQKGDVYSFAIVMHEIVNRQGVFWLGPGIEFSPKEIIETIRASCLRPNTSHHRSCEADDAAELMRRCWAEDPTERPDFGNLKGAIRRLNKTQESSNILDNLLSRMEQYANNLETLVSERTQDYLEEKKKCEELLYQLLPKSVASQLINGQSVVAETFEQVTIYFSDIVGFTALSASSTPMQVVDLLNDLYTCFDSIVENYDVYKVETIGDAYMVVSGLPERNGTRHACEVARMALALLDAVRRFRIRHRPMDQLRLRIGLHSGPCVAGVVGLKMPRYCLFGDTVNTASRMESNGEALKIHVSPDTKALLDTFGTFHLECRGEVAMKGKGILVTYWLLGEKTDPNAPKEKPSLVKNQTLSTESGSLSHLTPFRQRMDQNSRSSSRRGSGVFQQRSIDVTKLEKETQPLLQSLAIQRQLSDPTEPVANGIDNEESDKLSVTNSISLNVNSLSHNHQGHPRVKLKEANSINHKSKVGNNNWISKLPTASNSFDNGTKKASERLKDYSSVPLLSKADTPNDSIV
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Feature
chain Guanylate cyclase
Uniprot
A0A0M3VH92
Q9BPR0
A0A212FC80
A0A194PLI8
A0A2A4IYS3
A0A158P0M1
+ More
A0A0L7QKK7 A0A3L8DRY2 A0A195F401 K7J4N4 A0A1B6ME96 A0A1B6L105 A0A023F0L0 A0A224XIJ1 A0A1Q3G4K8 A0A2M4CVF3 A0A2M4CVA2 A0A2M3YZQ2 A0A151WKN4 Q7PTP9 A0A1Y9GKX4 A0A2M4B9V1 A0A2M4BA30 A0A2M4A0W1 A0A2M4B9R9 A0A2M4BA27 A0A2M4A6J1 A0A182V1G5 A0A2M4CIX2 A0A026WB13 Q174S2 A0A0P4ZZF4 A0A0N8DFN2 A0A0P5W112 A0A0P6GQV3 T1KF53 A0A0F7VJB8 A0A1B6JNQ5 A0A146M109 A0A0A9WYA7 E9H6U6 A0A0C9QYW7 A0A034VF88 A0A0K8VHK8 G1FKY1 W8B2G3 A0A0A1X716 A0A0F7VHC5 A0A2S2NHF4 A0A1B0AAF2 B4NBA6 A0A1A9YK51 A0A1A9UTM4 A0A1J1IEB7 A0A0L0CC40 A0A1I8MLJ8 A0A1J1IG90 B4JT47 Q296Q0 B4GEV1 A0A3B0JPZ6 B3M388 A0A0P4XYS6 Q9VF17 B4PRJ3 B4QXT4 B4HLG9 A0A1W4VFT6 B3P3W6 B4K5Q8 B4LX87 A0A210PTL8 V4AEX2 A0A1B0G6S3 V4AZ64 A0A0Q9WE85 A0A336LM52 A0A1S3IRF8 A0A0A1X5Z0 V4BNJ3 A0A1S4G740 A0A1I8PRN8 R7U0U2 A0A1W4VFG5 A0A0P9CAM4 K1QPS4
A0A0L7QKK7 A0A3L8DRY2 A0A195F401 K7J4N4 A0A1B6ME96 A0A1B6L105 A0A023F0L0 A0A224XIJ1 A0A1Q3G4K8 A0A2M4CVF3 A0A2M4CVA2 A0A2M3YZQ2 A0A151WKN4 Q7PTP9 A0A1Y9GKX4 A0A2M4B9V1 A0A2M4BA30 A0A2M4A0W1 A0A2M4B9R9 A0A2M4BA27 A0A2M4A6J1 A0A182V1G5 A0A2M4CIX2 A0A026WB13 Q174S2 A0A0P4ZZF4 A0A0N8DFN2 A0A0P5W112 A0A0P6GQV3 T1KF53 A0A0F7VJB8 A0A1B6JNQ5 A0A146M109 A0A0A9WYA7 E9H6U6 A0A0C9QYW7 A0A034VF88 A0A0K8VHK8 G1FKY1 W8B2G3 A0A0A1X716 A0A0F7VHC5 A0A2S2NHF4 A0A1B0AAF2 B4NBA6 A0A1A9YK51 A0A1A9UTM4 A0A1J1IEB7 A0A0L0CC40 A0A1I8MLJ8 A0A1J1IG90 B4JT47 Q296Q0 B4GEV1 A0A3B0JPZ6 B3M388 A0A0P4XYS6 Q9VF17 B4PRJ3 B4QXT4 B4HLG9 A0A1W4VFT6 B3P3W6 B4K5Q8 B4LX87 A0A210PTL8 V4AEX2 A0A1B0G6S3 V4AZ64 A0A0Q9WE85 A0A336LM52 A0A1S3IRF8 A0A0A1X5Z0 V4BNJ3 A0A1S4G740 A0A1I8PRN8 R7U0U2 A0A1W4VFG5 A0A0P9CAM4 K1QPS4
EC Number
4.6.1.2
Pubmed
11483433
22118469
26354079
21347285
30249741
20075255
+ More
25474469 12364791 14747013 17210077 24508170 17510324 26823975 25401762 21292972 25348373 24495485 25830018 17994087 26108605 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 28812685 23254933 22992520
25474469 12364791 14747013 17210077 24508170 17510324 26823975 25401762 21292972 25348373 24495485 25830018 17994087 26108605 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 28812685 23254933 22992520
EMBL
LC081227
BAS31078.1
AB047558
BAB32672.1
AGBW02009215
OWR51328.1
+ More
KQ459600 KPI94301.1 NWSH01005377 PCG64263.1 ADTU01000999 ADTU01001000 ADTU01001001 ADTU01001002 ADTU01001003 ADTU01001004 ADTU01001005 ADTU01001006 ADTU01001007 ADTU01001008 KQ414940 KOC59164.1 QOIP01000005 RLU22558.1 KQ981855 KYN34897.1 AAZX01000462 AAZX01000467 AAZX01003821 GEBQ01005731 JAT34246.1 GEBQ01022656 JAT17321.1 GBBI01003690 JAC15022.1 GFTR01008565 JAW07861.1 GFDL01000300 JAV34745.1 GGFL01005071 MBW69249.1 GGFL01005072 MBW69250.1 GGFM01000984 MBW21735.1 KQ983002 KYQ48418.1 AAAB01008794 EAA03567.6 APCN01002156 GGFJ01000665 MBW49806.1 GGFJ01000774 MBW49915.1 GGFK01001070 MBW34391.1 GGFJ01000664 MBW49805.1 GGFJ01000773 MBW49914.1 GGFK01002927 MBW36248.1 GGFL01001106 MBW65284.1 KK107293 EZA53250.1 CH477406 EAT41579.1 GDIP01206411 JAJ16991.1 GDIP01038391 GDIP01024551 JAM65324.1 GDIP01093526 JAM10189.1 GDIQ01040342 JAN54395.1 CAEY01000034 LN830672 CFW94181.1 GECU01006994 JAT00713.1 GDHC01005166 JAQ13463.1 GBHO01031198 JAG12406.1 GL732598 EFX72574.1 GBYB01005872 JAG75639.1 GAKP01016966 JAC41986.1 GDHF01014393 GDHF01013971 JAI37921.1 JAI38343.1 JN225448 AEL97641.1 GAMC01011230 JAB95325.1 GBXI01013066 GBXI01007611 JAD01226.1 JAD06681.1 LN830669 CFW94178.1 GGMR01004005 MBY16624.1 CH964232 EDW81070.1 CVRI01000047 CRK98555.1 JRES01000623 KNC29816.1 CRK98556.1 CH916373 EDV94937.1 CM000070 EAL28393.2 CH479182 EDW34136.1 OUUW01000008 SPP84277.1 CH902617 EDV43549.1 GDIP01235910 JAI87491.1 AE014297 AAF55244.2 CM000160 EDW97393.2 CM000364 EDX12758.1 CH480815 EDW41989.1 CH954181 EDV49072.1 CH933806 EDW15120.1 KRG01394.1 CH940650 EDW66739.1 KRF82885.1 NEDP02005508 OWF39796.1 KB200170 ESP02584.1 CCAG010013864 KB200718 ESP00406.1 KRF82888.1 UFQT01000054 SSX19066.1 GBXI01008204 JAD06088.1 KB202408 ESO90424.1 AMQN01002234 KB308725 ELT96795.1 KPU80309.1 JH815886 EKC33099.1
KQ459600 KPI94301.1 NWSH01005377 PCG64263.1 ADTU01000999 ADTU01001000 ADTU01001001 ADTU01001002 ADTU01001003 ADTU01001004 ADTU01001005 ADTU01001006 ADTU01001007 ADTU01001008 KQ414940 KOC59164.1 QOIP01000005 RLU22558.1 KQ981855 KYN34897.1 AAZX01000462 AAZX01000467 AAZX01003821 GEBQ01005731 JAT34246.1 GEBQ01022656 JAT17321.1 GBBI01003690 JAC15022.1 GFTR01008565 JAW07861.1 GFDL01000300 JAV34745.1 GGFL01005071 MBW69249.1 GGFL01005072 MBW69250.1 GGFM01000984 MBW21735.1 KQ983002 KYQ48418.1 AAAB01008794 EAA03567.6 APCN01002156 GGFJ01000665 MBW49806.1 GGFJ01000774 MBW49915.1 GGFK01001070 MBW34391.1 GGFJ01000664 MBW49805.1 GGFJ01000773 MBW49914.1 GGFK01002927 MBW36248.1 GGFL01001106 MBW65284.1 KK107293 EZA53250.1 CH477406 EAT41579.1 GDIP01206411 JAJ16991.1 GDIP01038391 GDIP01024551 JAM65324.1 GDIP01093526 JAM10189.1 GDIQ01040342 JAN54395.1 CAEY01000034 LN830672 CFW94181.1 GECU01006994 JAT00713.1 GDHC01005166 JAQ13463.1 GBHO01031198 JAG12406.1 GL732598 EFX72574.1 GBYB01005872 JAG75639.1 GAKP01016966 JAC41986.1 GDHF01014393 GDHF01013971 JAI37921.1 JAI38343.1 JN225448 AEL97641.1 GAMC01011230 JAB95325.1 GBXI01013066 GBXI01007611 JAD01226.1 JAD06681.1 LN830669 CFW94178.1 GGMR01004005 MBY16624.1 CH964232 EDW81070.1 CVRI01000047 CRK98555.1 JRES01000623 KNC29816.1 CRK98556.1 CH916373 EDV94937.1 CM000070 EAL28393.2 CH479182 EDW34136.1 OUUW01000008 SPP84277.1 CH902617 EDV43549.1 GDIP01235910 JAI87491.1 AE014297 AAF55244.2 CM000160 EDW97393.2 CM000364 EDX12758.1 CH480815 EDW41989.1 CH954181 EDV49072.1 CH933806 EDW15120.1 KRG01394.1 CH940650 EDW66739.1 KRF82885.1 NEDP02005508 OWF39796.1 KB200170 ESP02584.1 CCAG010013864 KB200718 ESP00406.1 KRF82888.1 UFQT01000054 SSX19066.1 GBXI01008204 JAD06088.1 KB202408 ESO90424.1 AMQN01002234 KB308725 ELT96795.1 KPU80309.1 JH815886 EKC33099.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000005205
UP000053825
UP000279307
+ More
UP000078541 UP000002358 UP000075809 UP000007062 UP000075840 UP000075903 UP000053097 UP000008820 UP000015104 UP000000305 UP000092445 UP000007798 UP000092443 UP000078200 UP000183832 UP000037069 UP000095301 UP000001070 UP000001819 UP000008744 UP000268350 UP000007801 UP000000803 UP000002282 UP000000304 UP000001292 UP000192221 UP000008711 UP000009192 UP000008792 UP000242188 UP000030746 UP000092444 UP000085678 UP000095300 UP000014760 UP000005408
UP000078541 UP000002358 UP000075809 UP000007062 UP000075840 UP000075903 UP000053097 UP000008820 UP000015104 UP000000305 UP000092445 UP000007798 UP000092443 UP000078200 UP000183832 UP000037069 UP000095301 UP000001070 UP000001819 UP000008744 UP000268350 UP000007801 UP000000803 UP000002282 UP000000304 UP000001292 UP000192221 UP000008711 UP000009192 UP000008792 UP000242188 UP000030746 UP000092444 UP000085678 UP000095300 UP000014760 UP000005408
Pfam
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR001170 ANPR/GUC
IPR001828 ANF_lig-bd_rcpt
IPR018297 A/G_cyclase_CS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR003593 AAA+_ATPase
IPR032410 MTABC_N
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR036640 ABC1_TM_sf
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR001170 ANPR/GUC
IPR001828 ANF_lig-bd_rcpt
IPR018297 A/G_cyclase_CS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR003593 AAA+_ATPase
IPR032410 MTABC_N
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR036640 ABC1_TM_sf
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
SUPFAM
Gene 3D
ProteinModelPortal
A0A0M3VH92
Q9BPR0
A0A212FC80
A0A194PLI8
A0A2A4IYS3
A0A158P0M1
+ More
A0A0L7QKK7 A0A3L8DRY2 A0A195F401 K7J4N4 A0A1B6ME96 A0A1B6L105 A0A023F0L0 A0A224XIJ1 A0A1Q3G4K8 A0A2M4CVF3 A0A2M4CVA2 A0A2M3YZQ2 A0A151WKN4 Q7PTP9 A0A1Y9GKX4 A0A2M4B9V1 A0A2M4BA30 A0A2M4A0W1 A0A2M4B9R9 A0A2M4BA27 A0A2M4A6J1 A0A182V1G5 A0A2M4CIX2 A0A026WB13 Q174S2 A0A0P4ZZF4 A0A0N8DFN2 A0A0P5W112 A0A0P6GQV3 T1KF53 A0A0F7VJB8 A0A1B6JNQ5 A0A146M109 A0A0A9WYA7 E9H6U6 A0A0C9QYW7 A0A034VF88 A0A0K8VHK8 G1FKY1 W8B2G3 A0A0A1X716 A0A0F7VHC5 A0A2S2NHF4 A0A1B0AAF2 B4NBA6 A0A1A9YK51 A0A1A9UTM4 A0A1J1IEB7 A0A0L0CC40 A0A1I8MLJ8 A0A1J1IG90 B4JT47 Q296Q0 B4GEV1 A0A3B0JPZ6 B3M388 A0A0P4XYS6 Q9VF17 B4PRJ3 B4QXT4 B4HLG9 A0A1W4VFT6 B3P3W6 B4K5Q8 B4LX87 A0A210PTL8 V4AEX2 A0A1B0G6S3 V4AZ64 A0A0Q9WE85 A0A336LM52 A0A1S3IRF8 A0A0A1X5Z0 V4BNJ3 A0A1S4G740 A0A1I8PRN8 R7U0U2 A0A1W4VFG5 A0A0P9CAM4 K1QPS4
A0A0L7QKK7 A0A3L8DRY2 A0A195F401 K7J4N4 A0A1B6ME96 A0A1B6L105 A0A023F0L0 A0A224XIJ1 A0A1Q3G4K8 A0A2M4CVF3 A0A2M4CVA2 A0A2M3YZQ2 A0A151WKN4 Q7PTP9 A0A1Y9GKX4 A0A2M4B9V1 A0A2M4BA30 A0A2M4A0W1 A0A2M4B9R9 A0A2M4BA27 A0A2M4A6J1 A0A182V1G5 A0A2M4CIX2 A0A026WB13 Q174S2 A0A0P4ZZF4 A0A0N8DFN2 A0A0P5W112 A0A0P6GQV3 T1KF53 A0A0F7VJB8 A0A1B6JNQ5 A0A146M109 A0A0A9WYA7 E9H6U6 A0A0C9QYW7 A0A034VF88 A0A0K8VHK8 G1FKY1 W8B2G3 A0A0A1X716 A0A0F7VHC5 A0A2S2NHF4 A0A1B0AAF2 B4NBA6 A0A1A9YK51 A0A1A9UTM4 A0A1J1IEB7 A0A0L0CC40 A0A1I8MLJ8 A0A1J1IG90 B4JT47 Q296Q0 B4GEV1 A0A3B0JPZ6 B3M388 A0A0P4XYS6 Q9VF17 B4PRJ3 B4QXT4 B4HLG9 A0A1W4VFT6 B3P3W6 B4K5Q8 B4LX87 A0A210PTL8 V4AEX2 A0A1B0G6S3 V4AZ64 A0A0Q9WE85 A0A336LM52 A0A1S3IRF8 A0A0A1X5Z0 V4BNJ3 A0A1S4G740 A0A1I8PRN8 R7U0U2 A0A1W4VFG5 A0A0P9CAM4 K1QPS4
PDB
3A3K
E-value=7.8552e-60,
Score=589
Ontologies
PATHWAY
GO
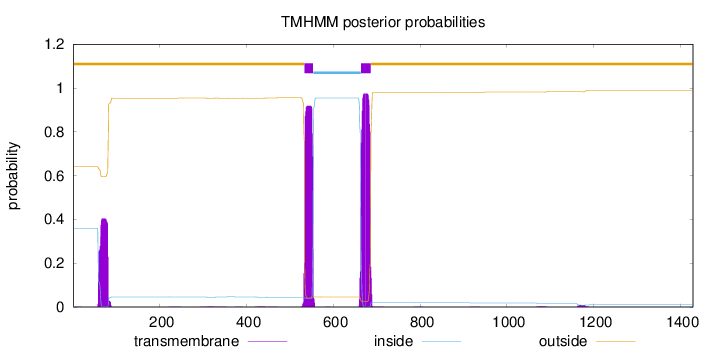
Topology
Length:
1429
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
49.8363900000003
Exp number, first 60 AAs:
0.69179
Total prob of N-in:
0.35933
outside
1 - 533
TMhelix
534 - 553
inside
554 - 663
TMhelix
664 - 686
outside
687 - 1429
Population Genetic Test Statistics
Pi
222.088659
Theta
167.452358
Tajima's D
1.129907
CLR
0.31382
CSRT
0.691665416729164
Interpretation
Uncertain
