Gene
KWMTBOMO04067
Pre Gene Modal
BGIBMGA010296
Annotation
olfactory_receptor_27_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 4.676
Sequence
CDS
ATGCCCAGTTCTTTTTTTCTGCCGAATTTGGAAAATCCCGATTATCCATCGTTAGGTCCGACTCTAAAGGGTCTAAAATATTGGGGTATGTGGCAATCGGGCGGGATTAAACGGATTTTGTACAATTCAATTCACGCATTCGCTACGTTTTTCGTGATCACCCAATACGTGGAGCTTTGGATTATCCGGAATAACGTCGAGCTAGCATTGAGGAACCTCTCTGTCACGATGCTTAGTACCGTTTGCGTCGTAAAAGCCGGTACTTTTGTCTGCTGGCAAAAATATTGGAGCGGCATTATCGGATTCGTCTCCAATTTAGAAAAGGAGCAACTGTCGAAAAATGATGCCGCCACGCAGGCCGCTATCGTCAAGTACATCAAATATTCACGCCGGGTCACCTATTTTTATTGGAGTTTGGTCACAGCCACTGTTTTTACAGTGATTTTAGCGCCACTGGTTGGTTTTTTGTCATCACCAGAACGTGAATTGATAGCGAACGGTACTTTACCATATCCTGAAATAATGAGCTCATGGGTACCGTTCGATAGATCGAGAGGCTTCGGTTATTGGGTAACGGCTTTAGTGCACACTTTGATCTGTTTCTACGGAGGCGGTGTGGTTGCGAACTACGACTCTAACGCGGTTGTTTTAATGTCGTTTTTCGCTGGACAAATGAAGCTTTTATCTATAAACTGTTCGAGATTGTTCGATGATGGTAACGAGGTCATTAGTAACAATGAAGCCATGAAGAGAATTAAGGAATGCCATTATCACCACGTGTATTTAGTGAGGTTTTCCACCATTTTTAACTCACTGATGTCACCAGTCTTGTTTTTATACGTTATAATATGTTCGCTCATGCTTTGTGCGAGTGCTGTGCAATTGACGACGGACGGAACTTCAAACATGCAGCGTATTTGGATTTCTGAATACCTTATGGCTTTAATAGCGCAGCTTTTCCTGTACTGTTGGCACAGCAACCAGGTTCTATATATGATGTACTGGAAGCTATCTAATAAATAA
Protein
MPSSFFLPNLENPDYPSLGPTLKGLKYWGMWQSGGIKRILYNSIHAFATFFVITQYVELWIIRNNVELALRNLSVTMLSTVCVVKAGTFVCWQKYWSGIIGFVSNLEKEQLSKNDAATQAAIVKYIKYSRRVTYFYWSLVTATVFTVILAPLVGFLSSPERELIANGTLPYPEIMSSWVPFDRSRGFGYWVTALVHTLICFYGGGVVANYDSNAVVLMSFFAGQMKLLSINCSRLFDDGNEVISNNEAMKRIKECHYHHVYLVRFSTIFNSLMSPVLFLYVIICSLMLCASAVQLTTDGTSNMQRIWISEYLMALIAQLFLYCWHSNQVLYMMYWKLSNK
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
C4B7V8
H9JL95
A7E3G9
A0A345BEV3
A0A1S5XXN4
A0A223HD09
+ More
A0A0B5CMZ5 A0A0B5GI82 A0A075TD69 A0A2A4IZ79 A0A0E4B5A3 A0A097ITU2 A0A223HD67 A0A223HDD9 A0A0K8TUU9 A0A097IYK8 A0A1B3B777 A0A2H1V744 H9A5P5 A0A1B3P5P8 A0A2L1TG50 A0A0L7L9K8 A0A3G6V6Q2 A0A212FBK8 A0A0S1TRD4 A0A0B5A3C7 A0A2K5B187 A0A2K8GKV8 A0A0K8TUT9 A0A1B3P5T6 A0A0B5GPZ8 A0A097ITT3 A0A097IYJ6 A0A194RLU9 A0A345BEU4 A0A223HD07 H9A5P6 A0A223HD55 A0A2H1V7A4 A0A345BEV5 A0A0B5CSR4 A0A194PZB7 A0A0S1TPY1 A0A2L1TG81 A0A1B3B787 A0A0E4B3V1 A0A1B2G2M1 A0A1S5XXN6 A0A1S5XXP3 A0A1V1WC62 A0A075T7Z4 A0A223HDE1 C4B7Y1 A0A0E4FIB5 A0A0V0J152 A0A223HD11 A0A212FK07 A0A3G6V734 A0A223HD50 A0A2L1TG52 A0A075T6X4 A0A1B3P5P2 A0A097IYL5 C4B7Y4 A0A0V0J105 A0A1B3B784 A0A1X9PB04 A0A0N1IQ17 A0A2K8GL40 A0A0K8TU74 A0A212FBH9 A0A0B5GQ06 A0A2K8GKW0 A0A3G6V6U8 A0A223HD25 A0A0P1J4K6 A0A223HD66 U5NG08 A0A0S1TKZ3 A0A2H1WJJ5 A0A2K8GKU3 A0A076EDE7 A0A2L1TG61 A0A076E7L9 A0A345BEW9 A0A1B3B748 C4B7W0 A0A223HDF8 A0A0B5GPZ4 A0A1Y9TK86 A0A0S1TPT8 A0A0B5CSS6 A0A1X9PB51 A0A0L7L3G4 A0A0P1IVM5 A0A097ITT4 A0A146JVN9 A0A097IYJ1 A0A1B3P5P4 A0A0K8TUA5
A0A0B5CMZ5 A0A0B5GI82 A0A075TD69 A0A2A4IZ79 A0A0E4B5A3 A0A097ITU2 A0A223HD67 A0A223HDD9 A0A0K8TUU9 A0A097IYK8 A0A1B3B777 A0A2H1V744 H9A5P5 A0A1B3P5P8 A0A2L1TG50 A0A0L7L9K8 A0A3G6V6Q2 A0A212FBK8 A0A0S1TRD4 A0A0B5A3C7 A0A2K5B187 A0A2K8GKV8 A0A0K8TUT9 A0A1B3P5T6 A0A0B5GPZ8 A0A097ITT3 A0A097IYJ6 A0A194RLU9 A0A345BEU4 A0A223HD07 H9A5P6 A0A223HD55 A0A2H1V7A4 A0A345BEV5 A0A0B5CSR4 A0A194PZB7 A0A0S1TPY1 A0A2L1TG81 A0A1B3B787 A0A0E4B3V1 A0A1B2G2M1 A0A1S5XXN6 A0A1S5XXP3 A0A1V1WC62 A0A075T7Z4 A0A223HDE1 C4B7Y1 A0A0E4FIB5 A0A0V0J152 A0A223HD11 A0A212FK07 A0A3G6V734 A0A223HD50 A0A2L1TG52 A0A075T6X4 A0A1B3P5P2 A0A097IYL5 C4B7Y4 A0A0V0J105 A0A1B3B784 A0A1X9PB04 A0A0N1IQ17 A0A2K8GL40 A0A0K8TU74 A0A212FBH9 A0A0B5GQ06 A0A2K8GKW0 A0A3G6V6U8 A0A223HD25 A0A0P1J4K6 A0A223HD66 U5NG08 A0A0S1TKZ3 A0A2H1WJJ5 A0A2K8GKU3 A0A076EDE7 A0A2L1TG61 A0A076E7L9 A0A345BEW9 A0A1B3B748 C4B7W0 A0A223HDF8 A0A0B5GPZ4 A0A1Y9TK86 A0A0S1TPT8 A0A0B5CSS6 A0A1X9PB51 A0A0L7L3G4 A0A0P1IVM5 A0A097ITT4 A0A146JVN9 A0A097IYJ1 A0A1B3P5P4 A0A0K8TUA5
Pubmed
EMBL
AB472107
BAH66324.1
BABH01011859
BK005934
DAA05984.1
MG816592
+ More
AXF48777.1 KY399276 AQQ73507.1 KY283603 AST36262.1 KM678307 AJE25880.1 KM892381 AJF23796.1 KF768692 AIG51886.1 NWSH01004938 PCG64574.1 LC002716 BAR43464.1 KM597208 AIT69885.1 KY283655 AST36314.1 KY283733 AST36392.1 GCVX01000184 JAI18046.1 KM655544 AIT71994.1 KT588136 AOE48046.1 ODYU01001027 SOQ36675.1 JN836695 AFC91735.2 KX655997 AOG12946.1 KY707212 AVF19654.1 JTDY01002113 KOB72079.1 MH193977 AZB49423.1 AGBW02009312 OWR51103.1 KR935724 ALM26216.1 KJ542690 AJD81574.1 KY081677 ASA39923.1 KX585361 ARO70254.1 GCVX01000172 JAI18058.1 KX656003 AOG12952.1 KM892372 AJF23787.1 KM597198 AIT69875.1 KM655534 AIT71984.1 KQ459995 KPJ18522.1 MG816583 AXF48768.1 KY283591 AST36251.1 JN836696 AFC91736.1 KY283644 AST36303.1 SOQ36676.1 MG816594 AXF48779.1 KM678298 AJE25871.1 KQ459586 KPI98084.1 KR935746 ALM26238.1 KY707231 AVF19673.1 KT588156 AOE48066.1 LC002707 BAR43455.1 KU598895 ANZ03141.1 KY399278 AQQ73509.1 KY399295 AQQ73526.1 GENK01000083 JAV45830.1 KF768693 AIG51887.1 KY283735 AST36394.1 AB472130 BAH66347.1 LC002706 BAR43454.1 GDKB01000047 JAP38449.1 KY283605 AST36264.1 AGBW02008175 OWR54056.1 MH193995 AZB49441.1 KY283657 AST36316.1 KY707201 AVF19643.1 KF768685 KZ149966 AIG51879.1 PZC76166.1 KX655986 AOG12935.1 KM655545 AIT71995.1 AB472133 BAH66350.1 GDKB01000054 JAP38442.1 KT588146 AOE48056.1 KX084464 ARO76419.1 KQ459900 KPJ19350.1 KY225500 ARO70512.1 GCVX01000186 JAI18044.1 OWR51104.1 KM892382 AJF23797.1 KX585359 ARO70252.1 MH193979 AZB49425.1 KY283616 AST36275.1 LN885112 CUQ99401.1 KY283668 AST36327.1 KC960479 AGY14590.1 KR935726 ALM26218.1 ODYU01009086 SOQ53253.1 KX585360 ARO70253.1 KF487667 AII01065.1 KY707211 AVF19653.1 KF487689 AII01087.1 MG816608 AXF48793.1 KT588106 AOE48016.1 AB472109 BAH66326.1 KY283744 AST36403.1 KM892367 AJF23782.1 KX084463 ARO76418.1 KR935748 ALM26240.1 KM678308 AJE25881.1 KX084504 ARO76459.1 JTDY01003257 KOB69861.1 LN885122 CUQ99411.1 KM597209 AIT69886.1 GEDO01000037 GEDO01000036 JAP88590.1 KM655529 AIT71979.1 KX655987 AOG12936.1 GCVX01000168 JAI18062.1
AXF48777.1 KY399276 AQQ73507.1 KY283603 AST36262.1 KM678307 AJE25880.1 KM892381 AJF23796.1 KF768692 AIG51886.1 NWSH01004938 PCG64574.1 LC002716 BAR43464.1 KM597208 AIT69885.1 KY283655 AST36314.1 KY283733 AST36392.1 GCVX01000184 JAI18046.1 KM655544 AIT71994.1 KT588136 AOE48046.1 ODYU01001027 SOQ36675.1 JN836695 AFC91735.2 KX655997 AOG12946.1 KY707212 AVF19654.1 JTDY01002113 KOB72079.1 MH193977 AZB49423.1 AGBW02009312 OWR51103.1 KR935724 ALM26216.1 KJ542690 AJD81574.1 KY081677 ASA39923.1 KX585361 ARO70254.1 GCVX01000172 JAI18058.1 KX656003 AOG12952.1 KM892372 AJF23787.1 KM597198 AIT69875.1 KM655534 AIT71984.1 KQ459995 KPJ18522.1 MG816583 AXF48768.1 KY283591 AST36251.1 JN836696 AFC91736.1 KY283644 AST36303.1 SOQ36676.1 MG816594 AXF48779.1 KM678298 AJE25871.1 KQ459586 KPI98084.1 KR935746 ALM26238.1 KY707231 AVF19673.1 KT588156 AOE48066.1 LC002707 BAR43455.1 KU598895 ANZ03141.1 KY399278 AQQ73509.1 KY399295 AQQ73526.1 GENK01000083 JAV45830.1 KF768693 AIG51887.1 KY283735 AST36394.1 AB472130 BAH66347.1 LC002706 BAR43454.1 GDKB01000047 JAP38449.1 KY283605 AST36264.1 AGBW02008175 OWR54056.1 MH193995 AZB49441.1 KY283657 AST36316.1 KY707201 AVF19643.1 KF768685 KZ149966 AIG51879.1 PZC76166.1 KX655986 AOG12935.1 KM655545 AIT71995.1 AB472133 BAH66350.1 GDKB01000054 JAP38442.1 KT588146 AOE48056.1 KX084464 ARO76419.1 KQ459900 KPJ19350.1 KY225500 ARO70512.1 GCVX01000186 JAI18044.1 OWR51104.1 KM892382 AJF23797.1 KX585359 ARO70252.1 MH193979 AZB49425.1 KY283616 AST36275.1 LN885112 CUQ99401.1 KY283668 AST36327.1 KC960479 AGY14590.1 KR935726 ALM26218.1 ODYU01009086 SOQ53253.1 KX585360 ARO70253.1 KF487667 AII01065.1 KY707211 AVF19653.1 KF487689 AII01087.1 MG816608 AXF48793.1 KT588106 AOE48016.1 AB472109 BAH66326.1 KY283744 AST36403.1 KM892367 AJF23782.1 KX084463 ARO76418.1 KR935748 ALM26240.1 KM678308 AJE25881.1 KX084504 ARO76459.1 JTDY01003257 KOB69861.1 LN885122 CUQ99411.1 KM597209 AIT69886.1 GEDO01000037 GEDO01000036 JAP88590.1 KM655529 AIT71979.1 KX655987 AOG12936.1 GCVX01000168 JAI18062.1
Proteomes
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
C4B7V8
H9JL95
A7E3G9
A0A345BEV3
A0A1S5XXN4
A0A223HD09
+ More
A0A0B5CMZ5 A0A0B5GI82 A0A075TD69 A0A2A4IZ79 A0A0E4B5A3 A0A097ITU2 A0A223HD67 A0A223HDD9 A0A0K8TUU9 A0A097IYK8 A0A1B3B777 A0A2H1V744 H9A5P5 A0A1B3P5P8 A0A2L1TG50 A0A0L7L9K8 A0A3G6V6Q2 A0A212FBK8 A0A0S1TRD4 A0A0B5A3C7 A0A2K5B187 A0A2K8GKV8 A0A0K8TUT9 A0A1B3P5T6 A0A0B5GPZ8 A0A097ITT3 A0A097IYJ6 A0A194RLU9 A0A345BEU4 A0A223HD07 H9A5P6 A0A223HD55 A0A2H1V7A4 A0A345BEV5 A0A0B5CSR4 A0A194PZB7 A0A0S1TPY1 A0A2L1TG81 A0A1B3B787 A0A0E4B3V1 A0A1B2G2M1 A0A1S5XXN6 A0A1S5XXP3 A0A1V1WC62 A0A075T7Z4 A0A223HDE1 C4B7Y1 A0A0E4FIB5 A0A0V0J152 A0A223HD11 A0A212FK07 A0A3G6V734 A0A223HD50 A0A2L1TG52 A0A075T6X4 A0A1B3P5P2 A0A097IYL5 C4B7Y4 A0A0V0J105 A0A1B3B784 A0A1X9PB04 A0A0N1IQ17 A0A2K8GL40 A0A0K8TU74 A0A212FBH9 A0A0B5GQ06 A0A2K8GKW0 A0A3G6V6U8 A0A223HD25 A0A0P1J4K6 A0A223HD66 U5NG08 A0A0S1TKZ3 A0A2H1WJJ5 A0A2K8GKU3 A0A076EDE7 A0A2L1TG61 A0A076E7L9 A0A345BEW9 A0A1B3B748 C4B7W0 A0A223HDF8 A0A0B5GPZ4 A0A1Y9TK86 A0A0S1TPT8 A0A0B5CSS6 A0A1X9PB51 A0A0L7L3G4 A0A0P1IVM5 A0A097ITT4 A0A146JVN9 A0A097IYJ1 A0A1B3P5P4 A0A0K8TUA5
A0A0B5CMZ5 A0A0B5GI82 A0A075TD69 A0A2A4IZ79 A0A0E4B5A3 A0A097ITU2 A0A223HD67 A0A223HDD9 A0A0K8TUU9 A0A097IYK8 A0A1B3B777 A0A2H1V744 H9A5P5 A0A1B3P5P8 A0A2L1TG50 A0A0L7L9K8 A0A3G6V6Q2 A0A212FBK8 A0A0S1TRD4 A0A0B5A3C7 A0A2K5B187 A0A2K8GKV8 A0A0K8TUT9 A0A1B3P5T6 A0A0B5GPZ8 A0A097ITT3 A0A097IYJ6 A0A194RLU9 A0A345BEU4 A0A223HD07 H9A5P6 A0A223HD55 A0A2H1V7A4 A0A345BEV5 A0A0B5CSR4 A0A194PZB7 A0A0S1TPY1 A0A2L1TG81 A0A1B3B787 A0A0E4B3V1 A0A1B2G2M1 A0A1S5XXN6 A0A1S5XXP3 A0A1V1WC62 A0A075T7Z4 A0A223HDE1 C4B7Y1 A0A0E4FIB5 A0A0V0J152 A0A223HD11 A0A212FK07 A0A3G6V734 A0A223HD50 A0A2L1TG52 A0A075T6X4 A0A1B3P5P2 A0A097IYL5 C4B7Y4 A0A0V0J105 A0A1B3B784 A0A1X9PB04 A0A0N1IQ17 A0A2K8GL40 A0A0K8TU74 A0A212FBH9 A0A0B5GQ06 A0A2K8GKW0 A0A3G6V6U8 A0A223HD25 A0A0P1J4K6 A0A223HD66 U5NG08 A0A0S1TKZ3 A0A2H1WJJ5 A0A2K8GKU3 A0A076EDE7 A0A2L1TG61 A0A076E7L9 A0A345BEW9 A0A1B3B748 C4B7W0 A0A223HDF8 A0A0B5GPZ4 A0A1Y9TK86 A0A0S1TPT8 A0A0B5CSS6 A0A1X9PB51 A0A0L7L3G4 A0A0P1IVM5 A0A097ITT4 A0A146JVN9 A0A097IYJ1 A0A1B3P5P4 A0A0K8TUA5
Ontologies
GO
PANTHER
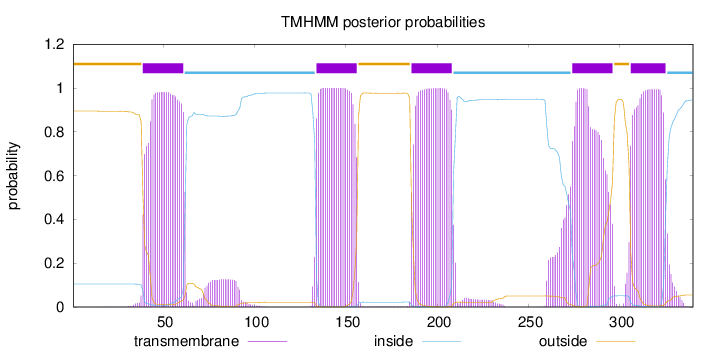
Topology
Subcellular location
Cell membrane
Length:
340
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.43183
Exp number, first 60 AAs:
20.02781
Total prob of N-in:
0.10513
POSSIBLE N-term signal
sequence
outside
1 - 38
TMhelix
39 - 61
inside
62 - 133
TMhelix
134 - 156
outside
157 - 185
TMhelix
186 - 208
inside
209 - 273
TMhelix
274 - 296
outside
297 - 305
TMhelix
306 - 325
inside
326 - 340
Population Genetic Test Statistics
Pi
291.466308
Theta
198.127643
Tajima's D
1.575083
CLR
0.222804
CSRT
0.798310084495775
Interpretation
Uncertain
