Gene
KWMTBOMO04064 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001051
Annotation
glutathione_S-transferase_epsilon_1_[Bombyx_mori]
Full name
Glutathione S-transferase 1
Alternative Name
GST class-theta
Location in the cell
Cytoplasmic Reliability : 3.113
Sequence
CDS
ATGGTGCTAACACTCTACAAGATGGACGCCAGTCCCCCAGTCCGCGCTGTCTACATGGTCATCGAAGCACTAAACATTCCCAACGTAAAATATGTAGACGTGGACCTCTTAGCTGAAGATCATTTGAAAGAGGAGTTCTTGAAGCTAAACCCTCAACACACCATTCCTATGCTGACAGATGACAAATTCGTCATCTGGGACAGTCATGCAATAGCTACATATTTGCTAAACAAATACGGCAAAGGCAGTTCATACTATCCCGAGGACCCAGAGAAGAGAGCCCTGATCGATATGAGATTACATTTCGACAGTGGAATATTGTATCCTGCTTTGCGAGAAAACGACGAGCCCATTTTCTTTTGGGGCGAGACCACCTTCAAGCCTCAAGGTCTGGCCAAAATAAAGTCAGCTTATGATTTCACGGAGAAGTTTCTTTCTGATTCACCGTGGATCGCAGGCGATGACGTCACTGTGGCTGATATGAGCTGTGTTGCTACAATCGGCAGTTTGGATGCGCTCTTGCCTATTAATGAGAAAGAGTATCCCAAGATAACTTCGTGGTTGAAACGCTGTTCTGAGCTAGACTTTTACCAGAGAGGAAACAACGTAAAAGGCCTATTGGAATTCAAGGCGTTACTGAAACAATACTTGAGTCGTGGAAAAGAATAA
Protein
MVLTLYKMDASPPVRAVYMVIEALNIPNVKYVDVDLLAEDHLKEEFLKLNPQHTIPMLTDDKFVIWDSHAIATYLLNKYGKGSSYYPEDPEKRALIDMRLHFDSGILYPALRENDEPIFFWGETTFKPQGLAKIKSAYDFTEKFLSDSPWIAGDDVTVADMSCVATIGSLDALLPINEKEYPKITSWLKRCSELDFYQRGNNVKGLLEFKALLKQYLSRGKE
Summary
Catalytic Activity
glutathione + RX = a halide anion + an S-substituted glutathione + H(+)
Subunit
Homodimer.
Similarity
Belongs to the GST superfamily.
Belongs to the GST superfamily. Theta family.
Belongs to the GST superfamily. Theta family.
Keywords
Transferase
Feature
chain Glutathione S-transferase 1
Uniprot
Q86PP4
A0A3B0ERN2
A0A3G1ZLC3
A0A075X2W2
A0A2A4JI76
A0A0D3M5T7
+ More
A0A3G1ZLC4 P46430 A0A075X3R8 A0A077DB43 A0A3G1RJ81 B0LKP4 A0A1W6I4K4 C8YL89 A0A3G1RJ71 A0A0K8TUZ9 A0A0K0XRM5 A0A0A7BYG1 A0A194R511 A0A212EWF1 A0A3S2LLI2 B0LB16 A0A2H1WMU8 A0A3G1ZLB5 H9JKP2 A0A212FPJ3 A0A075X250 A0A077D820 X5D048 A0A1P8L0S2 A0A194PT45 I4DRZ4 A0A1Y1K6Z7 H9JKP3 I4DQ12 B4J202 B4L947 B4LCA0 A0A1W4U4J0 A0A1L8EHK4 A0A1L8EHS3 A0A1Y1N675 A0A1Y1N945 B4P4E9 B4HNN1 A0A3B0JT17 A0A1A9Z7J5 B4NN03 D3TQP5 A1ZB72 B4HNN4 A0A1I8Q311 A0A0L0C9Y8 Q8MR33 B4NMZ5 B4P4F2 A0A0J9RFT5 B4L948 D6WFC4 A0A1I8P4U1 B4NN02 A0A1W4U585 D2D0D4 B4P4L9 B3NMS4 D6WCV8 B4QCE7 B4QCE3 A1ZB71 A0A1I8MAH9 A0A1A9VHV1 A0A3B0JRA5 B3NMS1 Q7JZM3 T1PG87 Q28YS6 A0A1I8MXR0 A0A3B0J7I3 A0A1I8MHG2 T1PDX5 B4P4F0 B4KM85 A0A1I8M6Z2 A0A1I8M6H7 B4GIX8 A0A1W4UG19 A0A139WLM6 B3NMS0 A0A1A9XX95 A0A0M5JBA5 A0A1Y1N4N8 B4HNN7 D6WCV7 B4HNN5 W8C6H4 Q9U796 A0A0M4RGU3 A0A0J9RFJ9 A0A1A9WI28
A0A3G1ZLC4 P46430 A0A075X3R8 A0A077DB43 A0A3G1RJ81 B0LKP4 A0A1W6I4K4 C8YL89 A0A3G1RJ71 A0A0K8TUZ9 A0A0K0XRM5 A0A0A7BYG1 A0A194R511 A0A212EWF1 A0A3S2LLI2 B0LB16 A0A2H1WMU8 A0A3G1ZLB5 H9JKP2 A0A212FPJ3 A0A075X250 A0A077D820 X5D048 A0A1P8L0S2 A0A194PT45 I4DRZ4 A0A1Y1K6Z7 H9JKP3 I4DQ12 B4J202 B4L947 B4LCA0 A0A1W4U4J0 A0A1L8EHK4 A0A1L8EHS3 A0A1Y1N675 A0A1Y1N945 B4P4E9 B4HNN1 A0A3B0JT17 A0A1A9Z7J5 B4NN03 D3TQP5 A1ZB72 B4HNN4 A0A1I8Q311 A0A0L0C9Y8 Q8MR33 B4NMZ5 B4P4F2 A0A0J9RFT5 B4L948 D6WFC4 A0A1I8P4U1 B4NN02 A0A1W4U585 D2D0D4 B4P4L9 B3NMS4 D6WCV8 B4QCE7 B4QCE3 A1ZB71 A0A1I8MAH9 A0A1A9VHV1 A0A3B0JRA5 B3NMS1 Q7JZM3 T1PG87 Q28YS6 A0A1I8MXR0 A0A3B0J7I3 A0A1I8MHG2 T1PDX5 B4P4F0 B4KM85 A0A1I8M6Z2 A0A1I8M6H7 B4GIX8 A0A1W4UG19 A0A139WLM6 B3NMS0 A0A1A9XX95 A0A0M5JBA5 A0A1Y1N4N8 B4HNN7 D6WCV7 B4HNN5 W8C6H4 Q9U796 A0A0M4RGU3 A0A0J9RFJ9 A0A1A9WI28
EC Number
2.5.1.18
Pubmed
EMBL
AY192575
DQ862465
AAO41719.1
ABI15774.1
RBVL01000267
RKO09898.1
+ More
MH177594 AYM01166.1 KF482966 AIH07590.1 NWSH01001309 PCG71767.1 KJ468614 AIB07715.1 MH177591 AYM01163.1 L32091 KF482963 AIH07587.1 KM099182 AIL29312.1 MG879008 AWX68888.1 EU216542 ABY66599.1 KX500032 ARM39002.1 FJ546089 ACU09495.1 MG879007 AWX68887.1 GCVX01000117 JAI18113.1 KP938862 AKS40343.1 KC966932 AHV85206.1 KQ460685 KPJ12893.1 AGBW02011991 OWR45830.1 RSAL01000067 RVE49301.1 EF506489 AB738325 ABS56977.1 BAM76808.1 ODYU01009688 SOQ54296.1 MH177592 AYM01164.1 BABH01011849 AGBW02002757 OWR55620.1 KF482964 AIH07588.1 KM099185 AIL29315.1 KF929205 AHW45903.1 KX229711 APW77572.1 KQ459600 KPI94305.1 AK405339 BAM20684.1 GEZM01090627 JAV57232.1 AK403881 BAM20002.1 CH916366 EDV95927.1 CH933816 EDW17222.1 CH940647 EDW68745.1 GFDG01000651 JAV18148.1 GFDG01000650 JAV18149.1 GEZM01012401 JAV92958.1 GEZM01013073 JAV92776.1 CM000158 EDW91635.1 CH480816 EDW48450.1 OUUW01000001 SPP75871.1 CH964282 EDW85742.1 EZ423747 ADD20023.1 AE013599 AAF57695.1 EDW48453.1 JRES01000678 KNC29243.1 AY122149 AAM52661.1 EDW85734.1 EDW91638.1 CM002911 KMY94817.1 EDW17223.2 KQ971319 EFA00271.1 EDW85741.1 FJ233082 ACO83224.1 EDW91642.1 CH954179 EDV55078.1 KQ971311 EEZ98834.1 CM000362 EDX07671.1 KMY94824.1 EDX07667.1 KMY94820.1 AAF57696.1 SPP75876.1 EDV55075.1 AY071166 AAL48788.2 KA646898 AFP61527.1 CM000071 EAL25890.1 SPP75872.1 KA646899 AFP61528.1 EDW91636.1 CH933808 EDW08749.1 CH479183 EDW36396.1 KYB28850.1 EDV55074.1 CP012525 ALC44418.1 GEZM01013079 JAV92769.1 EDW48456.1 EEZ98835.1 EDW48454.1 GAMC01008536 JAB98019.1 AF147205 AAD54937.1 KR107238 ALF04573.1 KMY94823.1
MH177594 AYM01166.1 KF482966 AIH07590.1 NWSH01001309 PCG71767.1 KJ468614 AIB07715.1 MH177591 AYM01163.1 L32091 KF482963 AIH07587.1 KM099182 AIL29312.1 MG879008 AWX68888.1 EU216542 ABY66599.1 KX500032 ARM39002.1 FJ546089 ACU09495.1 MG879007 AWX68887.1 GCVX01000117 JAI18113.1 KP938862 AKS40343.1 KC966932 AHV85206.1 KQ460685 KPJ12893.1 AGBW02011991 OWR45830.1 RSAL01000067 RVE49301.1 EF506489 AB738325 ABS56977.1 BAM76808.1 ODYU01009688 SOQ54296.1 MH177592 AYM01164.1 BABH01011849 AGBW02002757 OWR55620.1 KF482964 AIH07588.1 KM099185 AIL29315.1 KF929205 AHW45903.1 KX229711 APW77572.1 KQ459600 KPI94305.1 AK405339 BAM20684.1 GEZM01090627 JAV57232.1 AK403881 BAM20002.1 CH916366 EDV95927.1 CH933816 EDW17222.1 CH940647 EDW68745.1 GFDG01000651 JAV18148.1 GFDG01000650 JAV18149.1 GEZM01012401 JAV92958.1 GEZM01013073 JAV92776.1 CM000158 EDW91635.1 CH480816 EDW48450.1 OUUW01000001 SPP75871.1 CH964282 EDW85742.1 EZ423747 ADD20023.1 AE013599 AAF57695.1 EDW48453.1 JRES01000678 KNC29243.1 AY122149 AAM52661.1 EDW85734.1 EDW91638.1 CM002911 KMY94817.1 EDW17223.2 KQ971319 EFA00271.1 EDW85741.1 FJ233082 ACO83224.1 EDW91642.1 CH954179 EDV55078.1 KQ971311 EEZ98834.1 CM000362 EDX07671.1 KMY94824.1 EDX07667.1 KMY94820.1 AAF57696.1 SPP75876.1 EDV55075.1 AY071166 AAL48788.2 KA646898 AFP61527.1 CM000071 EAL25890.1 SPP75872.1 KA646899 AFP61528.1 EDW91636.1 CH933808 EDW08749.1 CH479183 EDW36396.1 KYB28850.1 EDV55074.1 CP012525 ALC44418.1 GEZM01013079 JAV92769.1 EDW48456.1 EEZ98835.1 EDW48454.1 GAMC01008536 JAB98019.1 AF147205 AAD54937.1 KR107238 ALF04573.1 KMY94823.1
Proteomes
UP000276569
UP000218220
UP000053240
UP000007151
UP000283053
UP000005204
+ More
UP000053268 UP000001070 UP000009192 UP000008792 UP000192221 UP000002282 UP000001292 UP000268350 UP000092445 UP000007798 UP000000803 UP000095300 UP000037069 UP000007266 UP000008711 UP000000304 UP000095301 UP000078200 UP000001819 UP000008744 UP000092443 UP000092553 UP000091820
UP000053268 UP000001070 UP000009192 UP000008792 UP000192221 UP000002282 UP000001292 UP000268350 UP000092445 UP000007798 UP000000803 UP000095300 UP000037069 UP000007266 UP000008711 UP000000304 UP000095301 UP000078200 UP000001819 UP000008744 UP000092443 UP000092553 UP000091820
Interpro
ProteinModelPortal
Q86PP4
A0A3B0ERN2
A0A3G1ZLC3
A0A075X2W2
A0A2A4JI76
A0A0D3M5T7
+ More
A0A3G1ZLC4 P46430 A0A075X3R8 A0A077DB43 A0A3G1RJ81 B0LKP4 A0A1W6I4K4 C8YL89 A0A3G1RJ71 A0A0K8TUZ9 A0A0K0XRM5 A0A0A7BYG1 A0A194R511 A0A212EWF1 A0A3S2LLI2 B0LB16 A0A2H1WMU8 A0A3G1ZLB5 H9JKP2 A0A212FPJ3 A0A075X250 A0A077D820 X5D048 A0A1P8L0S2 A0A194PT45 I4DRZ4 A0A1Y1K6Z7 H9JKP3 I4DQ12 B4J202 B4L947 B4LCA0 A0A1W4U4J0 A0A1L8EHK4 A0A1L8EHS3 A0A1Y1N675 A0A1Y1N945 B4P4E9 B4HNN1 A0A3B0JT17 A0A1A9Z7J5 B4NN03 D3TQP5 A1ZB72 B4HNN4 A0A1I8Q311 A0A0L0C9Y8 Q8MR33 B4NMZ5 B4P4F2 A0A0J9RFT5 B4L948 D6WFC4 A0A1I8P4U1 B4NN02 A0A1W4U585 D2D0D4 B4P4L9 B3NMS4 D6WCV8 B4QCE7 B4QCE3 A1ZB71 A0A1I8MAH9 A0A1A9VHV1 A0A3B0JRA5 B3NMS1 Q7JZM3 T1PG87 Q28YS6 A0A1I8MXR0 A0A3B0J7I3 A0A1I8MHG2 T1PDX5 B4P4F0 B4KM85 A0A1I8M6Z2 A0A1I8M6H7 B4GIX8 A0A1W4UG19 A0A139WLM6 B3NMS0 A0A1A9XX95 A0A0M5JBA5 A0A1Y1N4N8 B4HNN7 D6WCV7 B4HNN5 W8C6H4 Q9U796 A0A0M4RGU3 A0A0J9RFJ9 A0A1A9WI28
A0A3G1ZLC4 P46430 A0A075X3R8 A0A077DB43 A0A3G1RJ81 B0LKP4 A0A1W6I4K4 C8YL89 A0A3G1RJ71 A0A0K8TUZ9 A0A0K0XRM5 A0A0A7BYG1 A0A194R511 A0A212EWF1 A0A3S2LLI2 B0LB16 A0A2H1WMU8 A0A3G1ZLB5 H9JKP2 A0A212FPJ3 A0A075X250 A0A077D820 X5D048 A0A1P8L0S2 A0A194PT45 I4DRZ4 A0A1Y1K6Z7 H9JKP3 I4DQ12 B4J202 B4L947 B4LCA0 A0A1W4U4J0 A0A1L8EHK4 A0A1L8EHS3 A0A1Y1N675 A0A1Y1N945 B4P4E9 B4HNN1 A0A3B0JT17 A0A1A9Z7J5 B4NN03 D3TQP5 A1ZB72 B4HNN4 A0A1I8Q311 A0A0L0C9Y8 Q8MR33 B4NMZ5 B4P4F2 A0A0J9RFT5 B4L948 D6WFC4 A0A1I8P4U1 B4NN02 A0A1W4U585 D2D0D4 B4P4L9 B3NMS4 D6WCV8 B4QCE7 B4QCE3 A1ZB71 A0A1I8MAH9 A0A1A9VHV1 A0A3B0JRA5 B3NMS1 Q7JZM3 T1PG87 Q28YS6 A0A1I8MXR0 A0A3B0J7I3 A0A1I8MHG2 T1PDX5 B4P4F0 B4KM85 A0A1I8M6Z2 A0A1I8M6H7 B4GIX8 A0A1W4UG19 A0A139WLM6 B3NMS0 A0A1A9XX95 A0A0M5JBA5 A0A1Y1N4N8 B4HNN7 D6WCV7 B4HNN5 W8C6H4 Q9U796 A0A0M4RGU3 A0A0J9RFJ9 A0A1A9WI28
PDB
4PNG
E-value=9.4997e-49,
Score=485
Ontologies
PATHWAY
00480
Glutathione metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
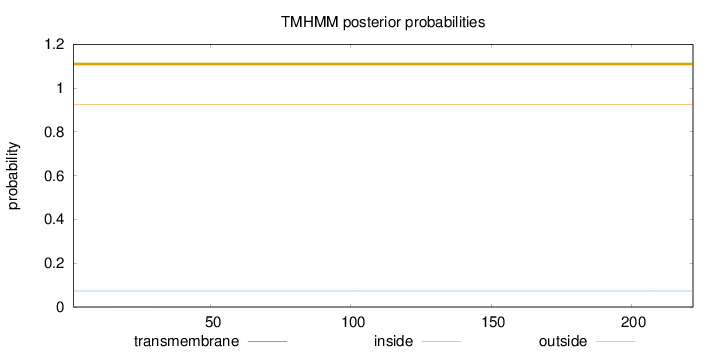
Topology
Length:
222
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00513
Exp number, first 60 AAs:
0.00282
Total prob of N-in:
0.07351
outside
1 - 222
Population Genetic Test Statistics
Pi
220.742089
Theta
166.181458
Tajima's D
1.045313
CLR
0.059696
CSRT
0.671566421678916
Interpretation
Uncertain
