Gene
KWMTBOMO04056
Annotation
reverse_ribonuclease_integrase?_partial_[Lasius_niger]
Location in the cell
Mitochondrial Reliability : 1.372 Nuclear Reliability : 1.425
Sequence
CDS
ATGGTTTTTACGATAAAATTTTTATCCCTACAAAAAACTGAACTGGAATACGAAGTTGCGATAAGAGGTGAGACCCCCGGTGCTACGGTGGCTGATCTTAGGAAACAGATTAGCAAATTAAGTCAATCTTTTCCTTCAGAAGATGTATTATGTTCTCATCTCGAACCCCTAGATGATCTTCAAGCTGTTTTGGACTTGCTGAATAAAGTCAATGCAAGTCTCGAGTCAAGCCCTACTGATAAAAGTACATTATCGCGCATAGAAAATATTTTTCATCACCTTTATCATAGGATCAACAGAATAGTGGTTGATGAAGTAGAGAAGCCGCACTCTGCTTGTAGTAAACTTTTTAAAGAATTCTACGAAAAATTCCAGAAATTAAGCAAAAAGGCAATTGATCCTCTATTAGTTGGTGTATCTAGTTCAGCATCTGCTTCTGTACCTGGCATTCAGGCACCAATTAATGTCGCTGTTACTTGTGATAGAGGTGTTACTTCAGAACTGAGTAGGCTGAAGTTCGATGTTTCCGGCCTTTTGGATTCAGGCTCAGCGGTTACTATTTTAGGCAACAATGCTCATAGTGCATTAGTTAGTCGTGGGCTGGAGTTGTGTACTGATGATAAATTGACTATTGTTGCGGCTGGTGGACAAAATCTTAGAAGTATAGGCTATATTAATATGCCAGTGCATTTCGAAAACCAGTTCCACATTATTCAGAGTCACGTAGTTCCAGAAGTTCAATCATCGCTTATTTTAGGGATTGACTTTTGGCACACATTTAAAATTTGCCCGAAGTACTTGAGTTCTCTTCATTTCCCCGGCCCTCCATTGGCTAATTTAGATGTTGGTAGCCCAACCAATTTTTTGCGCTCGTATGATTATTTGGACGAATCACAAAAGGCCGTCGTTGACAATATTACCAAGCAGTTTGAAGAAATATCTTTTGAGCGCAAAGGACTTGGACGCACAAACTTAATTACTCACCACATCGACACGGGGAATTCTCCTCCTGTGAGACAGCGCTACTATAGGCTTTCACCAGAGAAACAGAGAATCCTAGTCGAGCAAGTAGACGAAATGCTCCAGCTTGATGTAGTTGAGCCTTGTGAGAGTGCCTGGCAATCCCCGGTATTGCTAGTCACTAAAAAAGATGGTCAGCCACGGTTTTGCTTAGATAGTAGGAAGCTAAAGTCTGTTACAAAACGTGACGCTTATAACTTACCTTACATATCGGAGATATTGGACAACTTGCGAGACGCGCGCTATCTTAGCAGCATTGACTTATCAAAGGCTTTCTGGCAATTACCAATAGCACTCGAAGATAGGGACAAGACGGCATTTTACGTTCCGGGACGAGGGACGTTACGGTTCAAGACTACACCGTTTGGTTTGACCAATGCGCCTTCTACGCAACAGCGGTTAGTCGATTTATTGTTTGGCAACATGGATCACAAGGCGTTCGCATATTTAGATGATGTGATAATAGTAAGTAGCACGTTTGATGAGCATGTTAGTCTTCTTCTTCGCGTGTTAGACAAACTGCGCCATGCTAATCTAACAATTAATTTAGAAAAGTGTGAATTTTTTCGAAGTAAACTTAAATATTTAGGCTATGTGGTCGACGCGAATGGTTTGCGGACTGATCCGGATAAAGTTACAGCCATTCTAAATTACCCAACTCCTACATGTCGTAAGGACTTGAAACGGTTCCTAGGCACGGCAACATGGTACCGGAGATTTGTCCCAAACTTTAGCACTATCGCAGGGCCGCTTAATAGGTTGACATCGAACAAAAAATCAACACCTCCCTTCCAGTGGACAGATGAGGCCGACACTGCGTTCAAAAAACTTAAGGAATCTTTGGTCTCTGCACCTGTTCTCTCCTGCCCAAATTACGATCTGCCTTTCGAAGTCCATACAGATGCTAGTAACTATGGGATAGGTGGTATGCTCACACAAACTATTGACGGGAAAGAGCACCCTATAGCGTATATGAGCAAGTCCTTATCGGGGGCCGAAAAGAATTATAGCATTACCGAACGTGAGACCTTAGCTGTTCTCGTGGCTCTCGAGCACTGGCGTTGCTATCTGGAGAACGGAAAGACATTTACGGTTTACACAGACCATAGCGCACTTAAGTGGTTCCTTTCCTTGAACAATCCTACGGGTAGGCTTGCACGTTGGGGAGTTCGATTATCGTCGTTTGATTTTGAAATCAAACACCGCCGCGGTGTAGACAATGTGATTCCCGACGCTTTGTCTAGAGCTGTCCCGGTCACTACTATCACTACTGCTAAATCACTCACGACTTCTAGTGATGATTGAAAGCCTTTTGAATTTAATGTAGGAGATACTGTTTATAAACGCGCTTACGTCCTGAGTGACAAAGATAAGTATTTTAGTAAGAAGTTAGCACCGAAATATATCAAGTGCAAAGTTGTCGAAAAACTCTCGCCACTTGTCTACGTTCTAGAAGACATGTCGGGTAAAAACATAGGCACTTGGCATATTAAAGACTTGAAAATATTAGGTCAACACTCATAG
Protein
MVFTIKFLSLQKTELEYEVAIRGETPGATVADLRKQISKLSQSFPSEDVLCSHLEPLDDLQAVLDLLNKVNASLESSPTDKSTLSRIENIFHHLYHRINRIVVDEVEKPHSACSKLFKEFYEKFQKLSKKAIDPLLVGVSSSASASVPGIQAPINVAVTCDRGVTSELSRLKFDVSGLLDSGSAVTILGNNAHSALVSRGLELCTDDKLTIVAAGGQNLRSIGYINMPVHFENQFHIIQSHVVPEVQSSLILGIDFWHTFKICPKYLSSLHFPGPPLANLDVGSPTNFLRSYDYLDESQKAVVDNITKQFEEISFERKGLGRTNLITHHIDTGNSPPVRQRYYRLSPEKQRILVEQVDEMLQLDVVEPCESAWQSPVLLVTKKDGQPRFCLDSRKLKSVTKRDAYNLPYISEILDNLRDARYLSSIDLSKAFWQLPIALEDRDKTAFYVPGRGTLRFKTTPFGLTNAPSTQQRLVDLLFGNMDHKAFAYLDDVIIVSSTFDEHVSLLLRVLDKLRHANLTINLEKCEFFRSKLKYLGYVVDANGLRTDPDKVTAILNYPTPTCRKDLKRFLGTATWYRRFVPNFSTIAGPLNRLTSNKKSTPPFQWTDEADTAFKKLKESLVSAPVLSCPNYDLPFEVHTDASNYGIGGMLTQTIDGKEHPIAYMSKSLSGAEKNYSITERETLAVLVALEHWRCYLENGKTFTVYTDHSALKWFLSLNNPTGRLARWGVRLSSFDFEIKHRRGVDNVIPDALSRAVPVTTITTAKSLTTSSDDXKPFEFNVGDTVYKRAYVLSDKDKYFSKKLAPKYIKCKVVEKLSPLVYVLEDMSGKNIGTWHIKDLKILGQHS
Summary
Uniprot
Pubmed
EMBL
GEBQ01007184
GEBQ01002795
JAT32793.1
JAT37182.1
GEZM01099692
JAV53185.1
+ More
GEZM01046053 JAV77555.1 GEZM01099479 JAV53309.1 LN877232 CTR11712.1 GEZM01018686 JAV90062.1 GEZM01018688 GEZM01018687 GEZM01018685 GEZM01018684 GEZM01018683 GEZM01018682 JAV90067.1 GEHC01000926 JAV46719.1 JXUM01028079 KQ560809 KXJ80804.1 CH892681 KRK05548.1 GDHC01007507 JAQ11122.1 KQ972733 KXZ75794.1 GBBI01004634 JAC14078.1 AAGJ04130933 AAZX01023268
GEZM01046053 JAV77555.1 GEZM01099479 JAV53309.1 LN877232 CTR11712.1 GEZM01018686 JAV90062.1 GEZM01018688 GEZM01018687 GEZM01018685 GEZM01018684 GEZM01018683 GEZM01018682 JAV90067.1 GEHC01000926 JAV46719.1 JXUM01028079 KQ560809 KXJ80804.1 CH892681 KRK05548.1 GDHC01007507 JAQ11122.1 KQ972733 KXZ75794.1 GBBI01004634 JAC14078.1 AAGJ04130933 AAZX01023268
Proteomes
Pfam
Interpro
IPR041588
Integrase_H2C2
+ More
IPR036397 RNaseH_sf
IPR041373 RT_RNaseH
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR005162 Retrotrans_gag_dom
IPR018061 Retropepsins
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR003034 SAP_dom
IPR019103 Peptidase_aspartic_DDI1-type
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR003309 SCAN_dom
IPR019787 Znf_PHD-finger
IPR036397 RNaseH_sf
IPR041373 RT_RNaseH
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR005162 Retrotrans_gag_dom
IPR018061 Retropepsins
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR003034 SAP_dom
IPR019103 Peptidase_aspartic_DDI1-type
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR003309 SCAN_dom
IPR019787 Znf_PHD-finger
Gene 3D
ProteinModelPortal
PDB
4OL8
E-value=8.56346e-71,
Score=682
Ontologies
GO
Topology
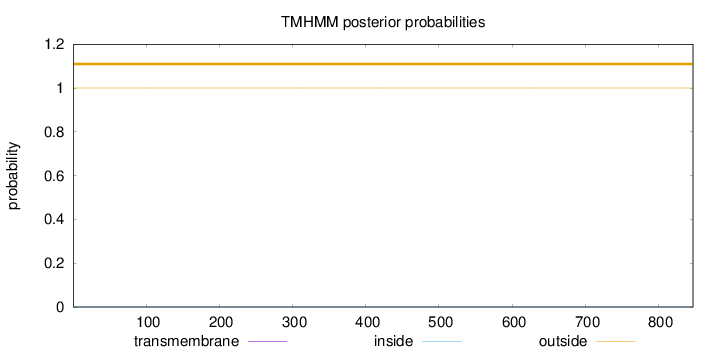
Length:
847
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00143
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00006
outside
1 - 847
Population Genetic Test Statistics
Pi
237.1813
Theta
145.203554
Tajima's D
2.493233
CLR
2.616135
CSRT
0.943102844857757
Interpretation
Uncertain
