Gene
KWMTBOMO04043
Pre Gene Modal
BGIBMGA010283
Annotation
septin_and_tuftelin_interacting_protein_[Bombyx_mori]
Full name
Septin-interacting protein 1
Alternative Name
Septin and tuftelin-interacting protein 1
Location in the cell
Cytoplasmic Reliability : 1.744 Nuclear Reliability : 2.224
Sequence
CDS
ATGTTTCCCGAAGACATACTCAAGGAGCCGGCGGTCCGCGACGCTTTCCGGAAAGCCCTCGACATGATGAACCGTAGCGCCGATATCGACACCATAGATCCGCCTCCTCCGCCAAGGTTCACCGTCATCACGGAGACCAAAGAAACGCCTAGAATTGTCGATGTATTAGCTTCGGCAACGCAACAGAAGAGTTTCTCCGAGTTGTTGGAGACCAGGTGCATTGAAAACGGTATAACTTTTGTACCGATTGCAGGGAAAACGAGGGAAGGTCGCCCCCTATATAAAATAGGTGCGCTACAGTGCTACGTTATAAGGAACGTGATAATGTATTCCGACGATAACGGAAGGAATTTTGCGCCAATCGCCATCGATAAGTTGCTCAAATTGGTCGAGGATTAG
Protein
MFPEDILKEPAVRDAFRKALDMMNRSADIDTIDPPPPPRFTVITETKETPRIVDVLASATQQKSFSELLETRCIENGITFVPIAGKTREGRPLYKIGALQCYVIRNVIMYSDDNGRNFAPIAIDKLLKLVED
Summary
Description
May be involved in pre-mRNA splicing.
Subunit
Identified in the spliceosome C complex (By similarity). Interacts with pnut.
Similarity
Belongs to the TFP11/STIP family.
Keywords
Coiled coil
Complete proteome
mRNA processing
mRNA splicing
Nucleus
Phosphoprotein
Reference proteome
Spliceosome
Feature
chain Septin-interacting protein 1
Uniprot
A1XDB3
H9JL82
A0A2W1AYN3
A0A3S2NTK0
A0A2W1BUW5
A0A2A4K201
+ More
A0A212FCY7 A0A1E1W018 A0A2H1WJM9 A0A194RKK6 A0A0L7LDN3 A0A194PZH2 W8BBQ4 A0A0K8VF45 A0A034WBN6 V5IAZ3 A0A336M495 N6SSW4 A0A1B6CZ51 B3MML6 A0A182GJF2 B4JC62 B4KGZ0 Q29KP1 A1XDA9 A0A1A9USN8 A0A1B0G097 B4MV34 U4U4Z9 A0A3B0JVD0 A0A0L0BWZ6 A0A1B0A3T9 A0A1B6CJS3 A0A1B6D3D3 Q16YS1 A1XDB4 A0A1S4G738 A0A1A9X057 B4LRI9 A0A1W4WLB1 A0A182PIZ5 A0A1Y1LAG3 E2ABM3 A0A2P8ZGS9 A0A182VTE4 A1XDB2 R7UZW4 A0A1Y1LHY9 A0A1I8MKJ6 A0A182KWD4 E2BJK8 A0A182IBE0 A0A0M9AA34 A0A023F277 A0A0M4E004 W5JQZ1 A0A0M4E4I9 X2J8R0 Q9Y103 A0A1Y1LAF4 A0A182X6B7 A1XDA8 A0A182Q9K9 A0A182F3B0 A0A1W4V391 A0A182JXN2 B3N567 A0A1Y1LAH8 A0A182LTI5 A0A232F2X2 A0A1B0GIT1 A0A182NAK3 A0A0J9QWB0 A0A182TUL0 B4Q417 E9J498 K7J8F2 A1XDB1 A0A084VME3 A0A067R831 A0A182VK21 A1XDB5 A0A1B6MTJ4 A0A151J015 F4WE40 A0A195BBD2 A0A158NUU0 A0A151K073 A0A182R3Y2 A0A1B0D5Y9 A0A1B6GBT5 A0A1B6GK58 A0A1B6GH69 A0A1B6IUK7 A0A0C9QGZ0 T1IB51 A0A1I8PX56 A0A182SHH9 A0A183IW63 A0A1J1I0P0
A0A212FCY7 A0A1E1W018 A0A2H1WJM9 A0A194RKK6 A0A0L7LDN3 A0A194PZH2 W8BBQ4 A0A0K8VF45 A0A034WBN6 V5IAZ3 A0A336M495 N6SSW4 A0A1B6CZ51 B3MML6 A0A182GJF2 B4JC62 B4KGZ0 Q29KP1 A1XDA9 A0A1A9USN8 A0A1B0G097 B4MV34 U4U4Z9 A0A3B0JVD0 A0A0L0BWZ6 A0A1B0A3T9 A0A1B6CJS3 A0A1B6D3D3 Q16YS1 A1XDB4 A0A1S4G738 A0A1A9X057 B4LRI9 A0A1W4WLB1 A0A182PIZ5 A0A1Y1LAG3 E2ABM3 A0A2P8ZGS9 A0A182VTE4 A1XDB2 R7UZW4 A0A1Y1LHY9 A0A1I8MKJ6 A0A182KWD4 E2BJK8 A0A182IBE0 A0A0M9AA34 A0A023F277 A0A0M4E004 W5JQZ1 A0A0M4E4I9 X2J8R0 Q9Y103 A0A1Y1LAF4 A0A182X6B7 A1XDA8 A0A182Q9K9 A0A182F3B0 A0A1W4V391 A0A182JXN2 B3N567 A0A1Y1LAH8 A0A182LTI5 A0A232F2X2 A0A1B0GIT1 A0A182NAK3 A0A0J9QWB0 A0A182TUL0 B4Q417 E9J498 K7J8F2 A1XDB1 A0A084VME3 A0A067R831 A0A182VK21 A1XDB5 A0A1B6MTJ4 A0A151J015 F4WE40 A0A195BBD2 A0A158NUU0 A0A151K073 A0A182R3Y2 A0A1B0D5Y9 A0A1B6GBT5 A0A1B6GK58 A0A1B6GH69 A0A1B6IUK7 A0A0C9QGZ0 T1IB51 A0A1I8PX56 A0A182SHH9 A0A183IW63 A0A1J1I0P0
Pubmed
17289020
19121390
28756777
22118469
26354079
26227816
+ More
24495485 25348373 23537049 17994087 26483478 15632085 23185243 26108605 17510324 28004739 20798317 29403074 17550304 23254933 25315136 20966253 25474469 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11884525 12537569 18327897 28648823 22936249 21282665 20075255 24438588 24845553 12364791 21719571 21347285
24495485 25348373 23537049 17994087 26483478 15632085 23185243 26108605 17510324 28004739 20798317 29403074 17550304 23254933 25315136 20966253 25474469 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11884525 12537569 18327897 28648823 22936249 21282665 20075255 24438588 24845553 12364791 21719571 21347285
EMBL
DQ342046
ABC69938.1
BABH01011772
BABH01011773
KZ150716
PZC70362.1
+ More
RSAL01000210 RVE44210.1 KZ149930 PZC77424.1 NWSH01000282 PCG77793.1 AGBW02009118 OWR51615.1 GDQN01010727 JAT80327.1 ODYU01009050 SOQ53176.1 KQ460075 KPJ17954.1 JTDY01001543 KOB73587.1 KQ459582 KPI98721.1 GAMC01007895 JAB98660.1 GDHF01014831 JAI37483.1 GAKP01005951 JAC53001.1 GALX01000335 JAB68131.1 UFQS01000144 UFQT01000144 SSX00438.1 SSX20818.1 APGK01057945 KB741283 ENN70774.1 GEDC01018745 JAS18553.1 CH902620 EDV31907.1 JXUM01012806 KQ560362 KXJ82820.1 CH916368 EDW03075.1 CH933807 EDW12201.1 DQ342043 CH379061 ABC69935.1 EAL33133.2 DQ342042 CH479189 ABC69934.1 EDW25521.1 CCAG010018487 CH963857 EDW76379.2 KB631738 ERL85681.1 OUUW01000010 SPP86034.1 JRES01001211 KNC24562.1 GEDC01023562 JAS13736.1 GEDC01017110 JAS20188.1 CH477510 EAT39778.1 DQ342047 ABC69939.1 CH940649 EDW63584.1 GEZM01061222 GEZM01061220 GEZM01061219 GEZM01061218 GEZM01061216 GEZM01061215 JAV70642.1 GL438317 EFN69166.1 PYGN01000062 PSN55670.1 DQ342045 CM000157 ABC69937.1 EDW88899.1 AMQN01005636 KB296425 ELU11797.1 GEZM01061225 GEZM01061221 GEZM01061214 JAV70637.1 GL448571 EFN84165.1 APCN01000672 KQ435704 KOX80307.1 GBBI01003521 JAC15191.1 CP012523 ALC38304.1 ADMH02000364 ETN66817.1 ALC38796.1 AE014134 AHN54151.1 AF221101 AF145670 GEZM01061224 GEZM01061217 JAV70632.1 DQ342041 ABC69933.1 AXCN02002191 CH954177 EDV57897.1 GEZM01061223 JAV70633.1 AXCM01004755 NNAY01001130 OXU25035.1 AJWK01015376 CM002910 KMY88357.1 CM000361 EDX03850.1 GL768088 EFZ12353.1 DQ342044 CH480820 ABC69936.1 EDW54403.1 ATLV01014586 KE524975 KFB39137.1 KK852872 KDR14564.1 AAAB01008944 DQ342048 HACL01000127 ABC69940.1 CFW94421.1 GEBQ01000762 JAT39215.1 KQ980670 KYN14624.1 GL888102 EGI67478.1 KQ976532 KYM81525.1 ADTU01026715 KQ981296 KYN43118.1 AJVK01003470 GECZ01009861 JAS59908.1 GECZ01006933 JAS62836.1 GECZ01008655 GECZ01008002 JAS61114.1 JAS61767.1 GECU01017098 JAS90608.1 GBYB01002744 JAG72511.1 ACPB03007249 UZAM01011012 VDP14527.1 CVRI01000038 CRK93763.1
RSAL01000210 RVE44210.1 KZ149930 PZC77424.1 NWSH01000282 PCG77793.1 AGBW02009118 OWR51615.1 GDQN01010727 JAT80327.1 ODYU01009050 SOQ53176.1 KQ460075 KPJ17954.1 JTDY01001543 KOB73587.1 KQ459582 KPI98721.1 GAMC01007895 JAB98660.1 GDHF01014831 JAI37483.1 GAKP01005951 JAC53001.1 GALX01000335 JAB68131.1 UFQS01000144 UFQT01000144 SSX00438.1 SSX20818.1 APGK01057945 KB741283 ENN70774.1 GEDC01018745 JAS18553.1 CH902620 EDV31907.1 JXUM01012806 KQ560362 KXJ82820.1 CH916368 EDW03075.1 CH933807 EDW12201.1 DQ342043 CH379061 ABC69935.1 EAL33133.2 DQ342042 CH479189 ABC69934.1 EDW25521.1 CCAG010018487 CH963857 EDW76379.2 KB631738 ERL85681.1 OUUW01000010 SPP86034.1 JRES01001211 KNC24562.1 GEDC01023562 JAS13736.1 GEDC01017110 JAS20188.1 CH477510 EAT39778.1 DQ342047 ABC69939.1 CH940649 EDW63584.1 GEZM01061222 GEZM01061220 GEZM01061219 GEZM01061218 GEZM01061216 GEZM01061215 JAV70642.1 GL438317 EFN69166.1 PYGN01000062 PSN55670.1 DQ342045 CM000157 ABC69937.1 EDW88899.1 AMQN01005636 KB296425 ELU11797.1 GEZM01061225 GEZM01061221 GEZM01061214 JAV70637.1 GL448571 EFN84165.1 APCN01000672 KQ435704 KOX80307.1 GBBI01003521 JAC15191.1 CP012523 ALC38304.1 ADMH02000364 ETN66817.1 ALC38796.1 AE014134 AHN54151.1 AF221101 AF145670 GEZM01061224 GEZM01061217 JAV70632.1 DQ342041 ABC69933.1 AXCN02002191 CH954177 EDV57897.1 GEZM01061223 JAV70633.1 AXCM01004755 NNAY01001130 OXU25035.1 AJWK01015376 CM002910 KMY88357.1 CM000361 EDX03850.1 GL768088 EFZ12353.1 DQ342044 CH480820 ABC69936.1 EDW54403.1 ATLV01014586 KE524975 KFB39137.1 KK852872 KDR14564.1 AAAB01008944 DQ342048 HACL01000127 ABC69940.1 CFW94421.1 GEBQ01000762 JAT39215.1 KQ980670 KYN14624.1 GL888102 EGI67478.1 KQ976532 KYM81525.1 ADTU01026715 KQ981296 KYN43118.1 AJVK01003470 GECZ01009861 JAS59908.1 GECZ01006933 JAS62836.1 GECZ01008655 GECZ01008002 JAS61114.1 JAS61767.1 GECU01017098 JAS90608.1 GBYB01002744 JAG72511.1 ACPB03007249 UZAM01011012 VDP14527.1 CVRI01000038 CRK93763.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000037510
+ More
UP000053268 UP000019118 UP000007801 UP000069940 UP000249989 UP000001070 UP000009192 UP000001819 UP000008744 UP000078200 UP000092444 UP000007798 UP000030742 UP000268350 UP000037069 UP000092445 UP000008820 UP000091820 UP000008792 UP000192223 UP000075885 UP000000311 UP000245037 UP000075920 UP000002282 UP000014760 UP000095301 UP000075882 UP000008237 UP000075840 UP000053105 UP000092553 UP000000673 UP000000803 UP000076407 UP000005203 UP000075886 UP000069272 UP000192221 UP000075881 UP000008711 UP000075883 UP000215335 UP000092461 UP000075884 UP000075902 UP000000304 UP000002358 UP000001292 UP000030765 UP000027135 UP000075903 UP000078492 UP000007755 UP000078540 UP000005205 UP000078541 UP000075900 UP000092462 UP000015103 UP000095300 UP000075901 UP000050793 UP000183832
UP000053268 UP000019118 UP000007801 UP000069940 UP000249989 UP000001070 UP000009192 UP000001819 UP000008744 UP000078200 UP000092444 UP000007798 UP000030742 UP000268350 UP000037069 UP000092445 UP000008820 UP000091820 UP000008792 UP000192223 UP000075885 UP000000311 UP000245037 UP000075920 UP000002282 UP000014760 UP000095301 UP000075882 UP000008237 UP000075840 UP000053105 UP000092553 UP000000673 UP000000803 UP000076407 UP000005203 UP000075886 UP000069272 UP000192221 UP000075881 UP000008711 UP000075883 UP000215335 UP000092461 UP000075884 UP000075902 UP000000304 UP000002358 UP000001292 UP000030765 UP000027135 UP000075903 UP000078492 UP000007755 UP000078540 UP000005205 UP000078541 UP000075900 UP000092462 UP000015103 UP000095300 UP000075901 UP000050793 UP000183832
Interpro
Gene 3D
ProteinModelPortal
A1XDB3
H9JL82
A0A2W1AYN3
A0A3S2NTK0
A0A2W1BUW5
A0A2A4K201
+ More
A0A212FCY7 A0A1E1W018 A0A2H1WJM9 A0A194RKK6 A0A0L7LDN3 A0A194PZH2 W8BBQ4 A0A0K8VF45 A0A034WBN6 V5IAZ3 A0A336M495 N6SSW4 A0A1B6CZ51 B3MML6 A0A182GJF2 B4JC62 B4KGZ0 Q29KP1 A1XDA9 A0A1A9USN8 A0A1B0G097 B4MV34 U4U4Z9 A0A3B0JVD0 A0A0L0BWZ6 A0A1B0A3T9 A0A1B6CJS3 A0A1B6D3D3 Q16YS1 A1XDB4 A0A1S4G738 A0A1A9X057 B4LRI9 A0A1W4WLB1 A0A182PIZ5 A0A1Y1LAG3 E2ABM3 A0A2P8ZGS9 A0A182VTE4 A1XDB2 R7UZW4 A0A1Y1LHY9 A0A1I8MKJ6 A0A182KWD4 E2BJK8 A0A182IBE0 A0A0M9AA34 A0A023F277 A0A0M4E004 W5JQZ1 A0A0M4E4I9 X2J8R0 Q9Y103 A0A1Y1LAF4 A0A182X6B7 A1XDA8 A0A182Q9K9 A0A182F3B0 A0A1W4V391 A0A182JXN2 B3N567 A0A1Y1LAH8 A0A182LTI5 A0A232F2X2 A0A1B0GIT1 A0A182NAK3 A0A0J9QWB0 A0A182TUL0 B4Q417 E9J498 K7J8F2 A1XDB1 A0A084VME3 A0A067R831 A0A182VK21 A1XDB5 A0A1B6MTJ4 A0A151J015 F4WE40 A0A195BBD2 A0A158NUU0 A0A151K073 A0A182R3Y2 A0A1B0D5Y9 A0A1B6GBT5 A0A1B6GK58 A0A1B6GH69 A0A1B6IUK7 A0A0C9QGZ0 T1IB51 A0A1I8PX56 A0A182SHH9 A0A183IW63 A0A1J1I0P0
A0A212FCY7 A0A1E1W018 A0A2H1WJM9 A0A194RKK6 A0A0L7LDN3 A0A194PZH2 W8BBQ4 A0A0K8VF45 A0A034WBN6 V5IAZ3 A0A336M495 N6SSW4 A0A1B6CZ51 B3MML6 A0A182GJF2 B4JC62 B4KGZ0 Q29KP1 A1XDA9 A0A1A9USN8 A0A1B0G097 B4MV34 U4U4Z9 A0A3B0JVD0 A0A0L0BWZ6 A0A1B0A3T9 A0A1B6CJS3 A0A1B6D3D3 Q16YS1 A1XDB4 A0A1S4G738 A0A1A9X057 B4LRI9 A0A1W4WLB1 A0A182PIZ5 A0A1Y1LAG3 E2ABM3 A0A2P8ZGS9 A0A182VTE4 A1XDB2 R7UZW4 A0A1Y1LHY9 A0A1I8MKJ6 A0A182KWD4 E2BJK8 A0A182IBE0 A0A0M9AA34 A0A023F277 A0A0M4E004 W5JQZ1 A0A0M4E4I9 X2J8R0 Q9Y103 A0A1Y1LAF4 A0A182X6B7 A1XDA8 A0A182Q9K9 A0A182F3B0 A0A1W4V391 A0A182JXN2 B3N567 A0A1Y1LAH8 A0A182LTI5 A0A232F2X2 A0A1B0GIT1 A0A182NAK3 A0A0J9QWB0 A0A182TUL0 B4Q417 E9J498 K7J8F2 A1XDB1 A0A084VME3 A0A067R831 A0A182VK21 A1XDB5 A0A1B6MTJ4 A0A151J015 F4WE40 A0A195BBD2 A0A158NUU0 A0A151K073 A0A182R3Y2 A0A1B0D5Y9 A0A1B6GBT5 A0A1B6GK58 A0A1B6GH69 A0A1B6IUK7 A0A0C9QGZ0 T1IB51 A0A1I8PX56 A0A182SHH9 A0A183IW63 A0A1J1I0P0
Ontologies
GO
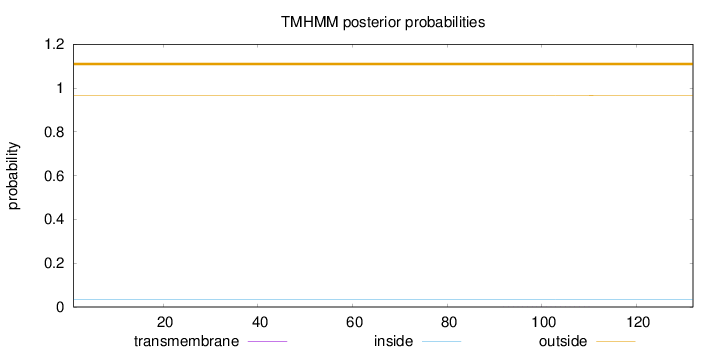
Topology
Subcellular location
Nucleus
Length:
132
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00626
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03327
outside
1 - 132
Population Genetic Test Statistics
Pi
340.950513
Theta
157.734524
Tajima's D
3.039931
CLR
0.396572
CSRT
0.980150992450378
Interpretation
Uncertain
