Gene
KWMTBOMO04023
Pre Gene Modal
BGIBMGA010268
Annotation
PREDICTED:_trafficking_protein_particle_complex_subunit_10_[Bombyx_mori]
Full name
Trafficking protein particle complex subunit 10
Alternative Name
Shal Interactor of Di-Leucine Motif
Trafficking protein particle complex subunit TMEM1
Transport protein particle subunit TMEM1
Trafficking protein particle complex subunit TMEM1
Transport protein particle subunit TMEM1
Location in the cell
Nuclear Reliability : 2.14
Sequence
CDS
ATGTGCACGGCAGCGCAAGCACCGCTCCTGCATCATGCCAAAGATAAGTTGCACGAATTGGGTAAGCTCTGTGGTCTACTCCCCGGTTCGCCGGAACCCACGTCAGAGCAACTCCATCTAGTCGTGATGCTGTCCGCGGGCATGGGGGACAGCGAACCCAACTCGCAGCCGCCCACGCCGACGGACAGGCTCAAGGAGGCCCTGTGCAGCGCTCCCTCGTTCCAGCAGTACTATCTGGAGCTGGCCGAGCTGGCGATGGGAACTTACAAGCACATAGGGAGGTTAAGATTCGCTCGGCTTATCGGTAGGGAGCTGGCATCGTTCTACTCCGAGTTGGGCGAGACCAGCAAGGCGGTGGTGTTCCTGATGGACGCGTTGAGGTCGTACGAGGAACAAGGCTGGAAGGAGCTGGCGGCACAAACGAGATTAGAATTGGTTTCGGCGGCGAAGAAAATGAAGGACATGGATAGATACGCGAAGCTCTCTGCAACAATAGCATGCACACAAGAATTAGAAATACTAGTCAGGAATTTTTACTTCGAGGAAATGATGAGGACCATACGTGAGGATCTGGCGAACAAAACCGACACGGTGCTGGCCGAATTGAACGAGTGCTTCAAGGTCAAGTCTGTCTCGTTACTGCCACCCGAAAGGGGTCTTTACATCACCGAGAATAAGGTTCAATGTCGTGTAGTCATAGAAAGTTTTCTACCTAAAGATATAAAATGTACGAAGGCAGCAATAAGCATAGAGTTATTTAAAGACGACAAGGTAGCGGAGAAGAAGCCTCCCGTCAAACACAAGACTGATTTAAGCGTTAACTGTAGTGTAGGAATACAGACCGGCTCGCCGAAGAGGAACGTCGATAAAATTGAAGATGCAATAAATAGTTTAGTTTGTAGCTTAAGACTCGATGATTTAAAACCCAACAATCCTTTACTAAACCCGTTACATATAACTTCGCAATTGCACTACAAAGAGGACAAGTCTTTACAAAAGGTGACCGTGGAATGTCACAACCCTAAAATGCCGCTGCGTCGTTCTGACAGTAGCAAATATCGGAAGCCGTCGGCGACGATCAGAAACAACTACCAGAATTGCTTTTCCAGTGAGGATATATTACTGAAACCTGGCTCCAACGACCTGATACTGGATTTTACAGCGACAGAAAAGGGCGCGTTTAAATTAGGTCAAATATCGATAACAGTGGAGGGGAAATTGGAGTTTTTATCGAACGCTTTGATTGGCGGTAAAGTCGGATTCGAAGTGGTAACTCAGGGGGTCAACGTTTATTTAAACAAAGTCGAACCTAAGAAGGATCTGGTCGCTGGCTTGGAGCACGCAATGGAGCTAGTTGTGACTTGTGGGAGTACCAGGATTGAAGAGGACTCGAAGATATCATTGAAGACGACATCTGGGTTGCTGATTCGGCTGTCGGATGAGTTCTCTATAATGTCTCGCGAGGTGTCGGTGCGGGTCGCGTGCGAACCCTTCGACACGGCTACGATACCGTTGACCCTGTTCGCCAGCCTTCAGCCCAGGAGGGAGAGGAGCGTCGAGCACGTCGTTTGGATAACGTGCCCCTGGTTGGAGTCGGCAATGGAAGTTCCTCTACACTTCTCCCCGCCCATGATCGCGTCGTGGCGCCTCCTCACTTCCAACACCCGGAAGTTCGTGCACGTGACTCTGAAGTCGACGGTCGCGCACTACGTCCAGTTCGCGCTCACCGATGCCGGCCTGCAGTGCGACGGTAACAACACCGTGTCGGATTTGAACCCGAAGAAGAGCGATCATATGTCTAGGCTGTCCAATTTGGCTGGTGGTATGTCCTCACGCGTGAGCACCATTCCTGTCGCGTAG
Protein
MCTAAQAPLLHHAKDKLHELGKLCGLLPGSPEPTSEQLHLVVMLSAGMGDSEPNSQPPTPTDRLKEALCSAPSFQQYYLELAELAMGTYKHIGRLRFARLIGRELASFYSELGETSKAVVFLMDALRSYEEQGWKELAAQTRLELVSAAKKMKDMDRYAKLSATIACTQELEILVRNFYFEEMMRTIREDLANKTDTVLAELNECFKVKSVSLLPPERGLYITENKVQCRVVIESFLPKDIKCTKAAISIELFKDDKVAEKKPPVKHKTDLSVNCSVGIQTGSPKRNVDKIEDAINSLVCSLRLDDLKPNNPLLNPLHITSQLHYKEDKSLQKVTVECHNPKMPLRRSDSSKYRKPSATIRNNYQNCFSSEDILLKPGSNDLILDFTATEKGAFKLGQISITVEGKLEFLSNALIGGKVGFEVVTQGVNVYLNKVEPKKDLVAGLEHAMELVVTCGSTRIEEDSKISLKTTSGLLIRLSDEFSIMSREVSVRVACEPFDTATIPLTLFASLQPRRERSVEHVVWITCPWLESAMEVPLHFSPPMIASWRLLTSNTRKFVHVTLKSTVAHYVQFALTDAGLQCDGNNTVSDLNPKKSDHMSRLSNLAGGMSSRVSTIPVA
Summary
Description
May play a role in vesicular transport from endoplasmic reticulum to Golgi (By similarity). Has a role in one of the several mechanisms underlying dendritic localization of Shal channels.
Subunit
Part of the multisubunit TRAPP (transport protein particle) complex (By similarity). Interacts with Shal (via C-terminal dendritic targeting motif).
Similarity
Belongs to the TMEM1 family.
Keywords
Cell projection
Complete proteome
ER-Golgi transport
Golgi apparatus
Reference proteome
Transport
Feature
chain Trafficking protein particle complex subunit 10
Uniprot
H9JL67
A0A2W1CNX6
A0A2A4JHN0
A0A2H1W0N4
A0A194PT11
A0A212EUC2
+ More
A0A194RMK7 A0A2J7QA27 A0A2P8YY38 A0A067QSA2 A0A154PH83 E2A0P2 F4W839 A0A195EZE1 A0A026WI98 E9J503 A0A3L8E0G8 A0A0N0BDV1 A0A0J7L374 A0A195CLG7 A0A088AJ40 A0A151X4M6 E2B8E7 A0A158NDR7 A0A195D7M5 A0A0L7R7Z9 A0A2A3E6L9 A0A310SEP3 A0A195BPE0 A0A232FMA8 K7ISG8 A0A1B6DTM9 A0A1B6CSP2 B0WLN5 Q16RX3 A0A1S4FRN1 A0A182G317 A0A1Q3F869 A0A3F2YX77 A0A182M9T0 A0A182VQ33 A0A2M4ABH4 A0A182RH39 A0A2M4ABR4 A0A2M4ABD4 A0A1B0DP71 A0A182NMP8 W5J2W7 A0A182FSX0 A0A2M4BAU0 A0A182PCR8 A0A2M4BBF5 A0A182JJA6 A0A2M4BAS7 A0A1B0CI91 A0A182JT61 A0A139WBM0 A0A182HZG9 A0A084W2N9 A0A182LQH9 A0A182V253 A0A182X391 Q7QKA8 A0A182U6Y6 A0A0T6AWT4 A0A1Y1M615 W8ASI2 W8BQL7 A0A1J1HYY0 A0A0K8VM86 A0A034VIE6 A0A336MVC5 A0A336MQ46 B3MXA2 A0A0N8NZX6 U4UAT2 N6UHW9 A0A1W4VZJ4 A0A1W4VN09 A0A3B0KC07 B4NBB3 Q294T9 A0A0R3NJN1 B4GMM1 A0A0R3NWS7 B5DUI6 A0A1I8N2Z6 A0A0L0C850 B4QZ42 A0A1I8PWT6 B4KAG4 A0A0Q9X157 T1PHI3 B4HE09 Q9VFB7 A0A0R1E828 B4PS80 B3P0R7 A0A023EZE9 B4JGB1
A0A194RMK7 A0A2J7QA27 A0A2P8YY38 A0A067QSA2 A0A154PH83 E2A0P2 F4W839 A0A195EZE1 A0A026WI98 E9J503 A0A3L8E0G8 A0A0N0BDV1 A0A0J7L374 A0A195CLG7 A0A088AJ40 A0A151X4M6 E2B8E7 A0A158NDR7 A0A195D7M5 A0A0L7R7Z9 A0A2A3E6L9 A0A310SEP3 A0A195BPE0 A0A232FMA8 K7ISG8 A0A1B6DTM9 A0A1B6CSP2 B0WLN5 Q16RX3 A0A1S4FRN1 A0A182G317 A0A1Q3F869 A0A3F2YX77 A0A182M9T0 A0A182VQ33 A0A2M4ABH4 A0A182RH39 A0A2M4ABR4 A0A2M4ABD4 A0A1B0DP71 A0A182NMP8 W5J2W7 A0A182FSX0 A0A2M4BAU0 A0A182PCR8 A0A2M4BBF5 A0A182JJA6 A0A2M4BAS7 A0A1B0CI91 A0A182JT61 A0A139WBM0 A0A182HZG9 A0A084W2N9 A0A182LQH9 A0A182V253 A0A182X391 Q7QKA8 A0A182U6Y6 A0A0T6AWT4 A0A1Y1M615 W8ASI2 W8BQL7 A0A1J1HYY0 A0A0K8VM86 A0A034VIE6 A0A336MVC5 A0A336MQ46 B3MXA2 A0A0N8NZX6 U4UAT2 N6UHW9 A0A1W4VZJ4 A0A1W4VN09 A0A3B0KC07 B4NBB3 Q294T9 A0A0R3NJN1 B4GMM1 A0A0R3NWS7 B5DUI6 A0A1I8N2Z6 A0A0L0C850 B4QZ42 A0A1I8PWT6 B4KAG4 A0A0Q9X157 T1PHI3 B4HE09 Q9VFB7 A0A0R1E828 B4PS80 B3P0R7 A0A023EZE9 B4JGB1
Pubmed
19121390
28756777
26354079
22118469
29403074
24845553
+ More
20798317 21719571 24508170 21282665 30249741 21347285 28648823 20075255 17510324 26483478 20920257 23761445 18362917 19820115 24438588 20966253 12364791 14747013 17210077 28004739 24495485 25348373 17994087 23537049 15632085 25315136 26108605 10731132 12537572 12537569 20550966 17550304 25474469
20798317 21719571 24508170 21282665 30249741 21347285 28648823 20075255 17510324 26483478 20920257 23761445 18362917 19820115 24438588 20966253 12364791 14747013 17210077 28004739 24495485 25348373 17994087 23537049 15632085 25315136 26108605 10731132 12537572 12537569 20550966 17550304 25474469
EMBL
BABH01011730
BABH01011731
BABH01011732
NHMN01022918
PZC87377.1
NWSH01001544
+ More
PCG70902.1 ODYU01005618 SOQ46649.1 KQ459600 KPI94270.1 AGBW02012412 OWR45095.1 KQ459995 KPJ18560.1 NEVH01016339 PNF25422.1 PYGN01000291 PSN49161.1 KK852994 KDR12739.1 KQ434899 KZC10834.1 GL435626 EFN72945.1 GL887888 EGI69709.1 KQ981906 KYN33277.1 KK107207 EZA55391.1 GL768123 EFZ12077.1 QOIP01000002 RLU26204.1 KQ435851 KOX70722.1 LBMM01000883 KMQ97292.1 KQ977600 KYN01543.1 KQ982548 KYQ55178.1 GL446312 EFN88046.1 ADTU01012806 KQ981153 KYN08876.1 KQ414637 KOC66958.1 KZ288368 PBC26809.1 KQ760629 OAD59730.1 KQ976424 KYM88375.1 NNAY01000019 OXU31886.1 GEDC01008264 GEDC01007513 JAS29034.1 JAS29785.1 GEDC01020911 JAS16387.1 DS231988 EDS30577.1 CH477694 EAT37176.1 JXUM01140583 KQ569087 KXJ68771.1 GFDL01011318 JAV23727.1 AXCN02001211 AXCM01000279 GGFK01004788 MBW38109.1 GGFK01004910 MBW38231.1 GGFK01004776 MBW38097.1 AJVK01007931 ADMH02002125 ETN58652.1 GGFJ01000999 MBW50140.1 GGFJ01000997 MBW50138.1 GGFJ01000998 MBW50139.1 AJWK01013105 AJWK01013106 KQ971372 KYB25316.1 APCN01001975 ATLV01019658 KE525276 KFB44483.1 AAAB01008799 EAA03661.5 LJIG01022634 KRT79522.1 GEZM01043057 JAV79346.1 GAMC01014735 JAB91820.1 GAMC01014733 GAMC01014731 JAB91824.1 CVRI01000030 CRK92532.1 GDHF01012325 JAI39989.1 GAKP01017634 JAC41318.1 UFQS01002542 UFQT01002542 SSX14240.1 SSX33655.1 UFQS01000939 UFQT01000939 SSX07864.1 SSX28098.1 CH902629 EDV35325.1 KPU75548.1 KB631985 ERL87716.1 APGK01019569 KB740112 ENN81340.1 OUUW01000008 SPP83689.1 CH964232 EDW81077.1 CM000070 EAL28875.2 KRT01145.1 CH479185 EDW38095.1 CH673789 KRT05559.1 EDY71552.1 JRES01000776 KNC28425.1 CM000364 EDX12870.1 CH933806 EDW15677.1 KRG01677.1 KA648282 AFP62911.1 CH480815 EDW42101.1 AE014297 AY118591 CM000160 KRK03778.1 EDW97500.1 CH954181 EDV48965.1 GBBI01004090 JAC14622.1 CH916369 EDV92580.1
PCG70902.1 ODYU01005618 SOQ46649.1 KQ459600 KPI94270.1 AGBW02012412 OWR45095.1 KQ459995 KPJ18560.1 NEVH01016339 PNF25422.1 PYGN01000291 PSN49161.1 KK852994 KDR12739.1 KQ434899 KZC10834.1 GL435626 EFN72945.1 GL887888 EGI69709.1 KQ981906 KYN33277.1 KK107207 EZA55391.1 GL768123 EFZ12077.1 QOIP01000002 RLU26204.1 KQ435851 KOX70722.1 LBMM01000883 KMQ97292.1 KQ977600 KYN01543.1 KQ982548 KYQ55178.1 GL446312 EFN88046.1 ADTU01012806 KQ981153 KYN08876.1 KQ414637 KOC66958.1 KZ288368 PBC26809.1 KQ760629 OAD59730.1 KQ976424 KYM88375.1 NNAY01000019 OXU31886.1 GEDC01008264 GEDC01007513 JAS29034.1 JAS29785.1 GEDC01020911 JAS16387.1 DS231988 EDS30577.1 CH477694 EAT37176.1 JXUM01140583 KQ569087 KXJ68771.1 GFDL01011318 JAV23727.1 AXCN02001211 AXCM01000279 GGFK01004788 MBW38109.1 GGFK01004910 MBW38231.1 GGFK01004776 MBW38097.1 AJVK01007931 ADMH02002125 ETN58652.1 GGFJ01000999 MBW50140.1 GGFJ01000997 MBW50138.1 GGFJ01000998 MBW50139.1 AJWK01013105 AJWK01013106 KQ971372 KYB25316.1 APCN01001975 ATLV01019658 KE525276 KFB44483.1 AAAB01008799 EAA03661.5 LJIG01022634 KRT79522.1 GEZM01043057 JAV79346.1 GAMC01014735 JAB91820.1 GAMC01014733 GAMC01014731 JAB91824.1 CVRI01000030 CRK92532.1 GDHF01012325 JAI39989.1 GAKP01017634 JAC41318.1 UFQS01002542 UFQT01002542 SSX14240.1 SSX33655.1 UFQS01000939 UFQT01000939 SSX07864.1 SSX28098.1 CH902629 EDV35325.1 KPU75548.1 KB631985 ERL87716.1 APGK01019569 KB740112 ENN81340.1 OUUW01000008 SPP83689.1 CH964232 EDW81077.1 CM000070 EAL28875.2 KRT01145.1 CH479185 EDW38095.1 CH673789 KRT05559.1 EDY71552.1 JRES01000776 KNC28425.1 CM000364 EDX12870.1 CH933806 EDW15677.1 KRG01677.1 KA648282 AFP62911.1 CH480815 EDW42101.1 AE014297 AY118591 CM000160 KRK03778.1 EDW97500.1 CH954181 EDV48965.1 GBBI01004090 JAC14622.1 CH916369 EDV92580.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000235965
+ More
UP000245037 UP000027135 UP000076502 UP000000311 UP000007755 UP000078541 UP000053097 UP000279307 UP000053105 UP000036403 UP000078542 UP000005203 UP000075809 UP000008237 UP000005205 UP000078492 UP000053825 UP000242457 UP000078540 UP000215335 UP000002358 UP000002320 UP000008820 UP000069940 UP000249989 UP000075886 UP000075883 UP000075920 UP000075900 UP000092462 UP000075884 UP000000673 UP000069272 UP000075885 UP000075880 UP000092461 UP000075881 UP000007266 UP000075840 UP000030765 UP000075882 UP000075903 UP000076407 UP000007062 UP000075902 UP000183832 UP000007801 UP000030742 UP000019118 UP000192221 UP000268350 UP000007798 UP000001819 UP000008744 UP000095301 UP000037069 UP000000304 UP000095300 UP000009192 UP000001292 UP000000803 UP000002282 UP000008711 UP000001070
UP000245037 UP000027135 UP000076502 UP000000311 UP000007755 UP000078541 UP000053097 UP000279307 UP000053105 UP000036403 UP000078542 UP000005203 UP000075809 UP000008237 UP000005205 UP000078492 UP000053825 UP000242457 UP000078540 UP000215335 UP000002358 UP000002320 UP000008820 UP000069940 UP000249989 UP000075886 UP000075883 UP000075920 UP000075900 UP000092462 UP000075884 UP000000673 UP000069272 UP000075885 UP000075880 UP000092461 UP000075881 UP000007266 UP000075840 UP000030765 UP000075882 UP000075903 UP000076407 UP000007062 UP000075902 UP000183832 UP000007801 UP000030742 UP000019118 UP000192221 UP000268350 UP000007798 UP000001819 UP000008744 UP000095301 UP000037069 UP000000304 UP000095300 UP000009192 UP000001292 UP000000803 UP000002282 UP000008711 UP000001070
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9JL67
A0A2W1CNX6
A0A2A4JHN0
A0A2H1W0N4
A0A194PT11
A0A212EUC2
+ More
A0A194RMK7 A0A2J7QA27 A0A2P8YY38 A0A067QSA2 A0A154PH83 E2A0P2 F4W839 A0A195EZE1 A0A026WI98 E9J503 A0A3L8E0G8 A0A0N0BDV1 A0A0J7L374 A0A195CLG7 A0A088AJ40 A0A151X4M6 E2B8E7 A0A158NDR7 A0A195D7M5 A0A0L7R7Z9 A0A2A3E6L9 A0A310SEP3 A0A195BPE0 A0A232FMA8 K7ISG8 A0A1B6DTM9 A0A1B6CSP2 B0WLN5 Q16RX3 A0A1S4FRN1 A0A182G317 A0A1Q3F869 A0A3F2YX77 A0A182M9T0 A0A182VQ33 A0A2M4ABH4 A0A182RH39 A0A2M4ABR4 A0A2M4ABD4 A0A1B0DP71 A0A182NMP8 W5J2W7 A0A182FSX0 A0A2M4BAU0 A0A182PCR8 A0A2M4BBF5 A0A182JJA6 A0A2M4BAS7 A0A1B0CI91 A0A182JT61 A0A139WBM0 A0A182HZG9 A0A084W2N9 A0A182LQH9 A0A182V253 A0A182X391 Q7QKA8 A0A182U6Y6 A0A0T6AWT4 A0A1Y1M615 W8ASI2 W8BQL7 A0A1J1HYY0 A0A0K8VM86 A0A034VIE6 A0A336MVC5 A0A336MQ46 B3MXA2 A0A0N8NZX6 U4UAT2 N6UHW9 A0A1W4VZJ4 A0A1W4VN09 A0A3B0KC07 B4NBB3 Q294T9 A0A0R3NJN1 B4GMM1 A0A0R3NWS7 B5DUI6 A0A1I8N2Z6 A0A0L0C850 B4QZ42 A0A1I8PWT6 B4KAG4 A0A0Q9X157 T1PHI3 B4HE09 Q9VFB7 A0A0R1E828 B4PS80 B3P0R7 A0A023EZE9 B4JGB1
A0A194RMK7 A0A2J7QA27 A0A2P8YY38 A0A067QSA2 A0A154PH83 E2A0P2 F4W839 A0A195EZE1 A0A026WI98 E9J503 A0A3L8E0G8 A0A0N0BDV1 A0A0J7L374 A0A195CLG7 A0A088AJ40 A0A151X4M6 E2B8E7 A0A158NDR7 A0A195D7M5 A0A0L7R7Z9 A0A2A3E6L9 A0A310SEP3 A0A195BPE0 A0A232FMA8 K7ISG8 A0A1B6DTM9 A0A1B6CSP2 B0WLN5 Q16RX3 A0A1S4FRN1 A0A182G317 A0A1Q3F869 A0A3F2YX77 A0A182M9T0 A0A182VQ33 A0A2M4ABH4 A0A182RH39 A0A2M4ABR4 A0A2M4ABD4 A0A1B0DP71 A0A182NMP8 W5J2W7 A0A182FSX0 A0A2M4BAU0 A0A182PCR8 A0A2M4BBF5 A0A182JJA6 A0A2M4BAS7 A0A1B0CI91 A0A182JT61 A0A139WBM0 A0A182HZG9 A0A084W2N9 A0A182LQH9 A0A182V253 A0A182X391 Q7QKA8 A0A182U6Y6 A0A0T6AWT4 A0A1Y1M615 W8ASI2 W8BQL7 A0A1J1HYY0 A0A0K8VM86 A0A034VIE6 A0A336MVC5 A0A336MQ46 B3MXA2 A0A0N8NZX6 U4UAT2 N6UHW9 A0A1W4VZJ4 A0A1W4VN09 A0A3B0KC07 B4NBB3 Q294T9 A0A0R3NJN1 B4GMM1 A0A0R3NWS7 B5DUI6 A0A1I8N2Z6 A0A0L0C850 B4QZ42 A0A1I8PWT6 B4KAG4 A0A0Q9X157 T1PHI3 B4HE09 Q9VFB7 A0A0R1E828 B4PS80 B3P0R7 A0A023EZE9 B4JGB1
Ontologies
GO
Topology
Subcellular location
Golgi apparatus
cis-Golgi network
Cell projection
Dendrite
Perikaryon
cis-Golgi network
Cell projection
Dendrite
Perikaryon
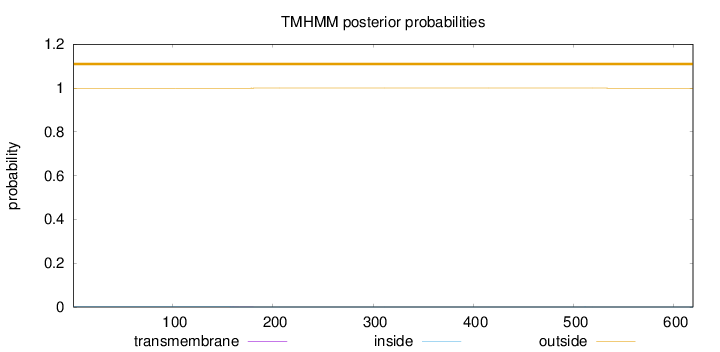
Length:
619
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01282
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00074
outside
1 - 619
Population Genetic Test Statistics
Pi
20.017106
Theta
2.192034
Tajima's D
0.542365
CLR
0.367958
CSRT
0.576921153942303
Interpretation
Uncertain
