Gene
KWMTBOMO04015
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.287 Nuclear Reliability : 1.079
Sequence
CDS
ATGACGCGGAGGCAGTTTGTAATCGGAACTACTCGAGATGAAACAGCGAACGGATCACCTAAAGCAAAGCGGAAGAAACGAATTACGAATTTTGGAACATTATCATGGTTCTGCCCTAGTGTGGTCAGAGCGCGGAGGGCCCAATCTCGGCGGTCCGTGCTGGAGTCATGGTCCAGACGGCTGGCCGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGGTTCTTGTGAATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGCTTCGGTGAGTTCCTGCACCGGATCGGAGCCGAGCTGACGGCAGAGTGCCACCATTGTGGTTGCGACTTGGACACGGCGGAGCACACGCTCGTCGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTTGTCGCAAAAATAGGAAACGACTTGTCGTTGCCGAGTGTTGTGGCATCAATGCTCGGTGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGCGAGACGCGCAGGCCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATTTCGCCCAAGCCCTGGCCCTCTAA
Protein
MTRRQFVIGTTRDETANGSPKAKRKKRITNFGTLSWFCPSVVRARRAQSRRSVLESWSRRLADPSAGRRTVEAIRPVLVNWVNRDRGRLTFRLTQVLTGHGCFGEFLHRIGAELTAECHHCGCDLDTAEHTLVACPAWEGWRRVLVAKIGNDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRVRDAQARRRRAGAREADFAQALAL
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
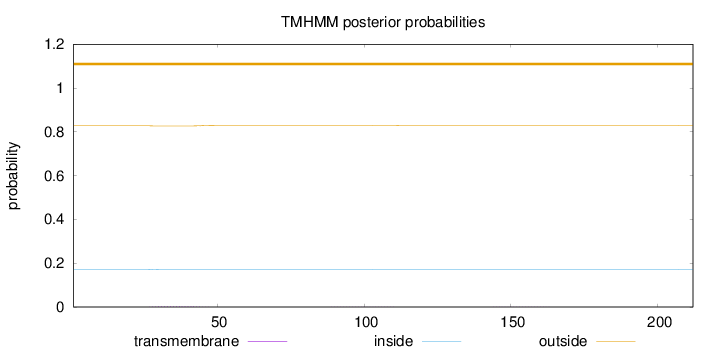
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0538600000000001
Exp number, first 60 AAs:
0.04485
Total prob of N-in:
0.17267
outside
1 - 212
Population Genetic Test Statistics
Pi
139.995176
Theta
72.610485
Tajima's D
1.799437
CLR
0.042221
CSRT
0.851557422128894
Interpretation
Uncertain
