Gene
KWMTBOMO04012
Pre Gene Modal
BGIBMGA010116
Annotation
PREDICTED:_uncharacterized_protein_LOC101736683_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.836
Sequence
CDS
ATGTATTTAACGTTGAAACAAAAGCTGTTAGAAGTTCCCGAAGGCACGGCGCAGCAGATACAAATGTTGTACGGTTACGATGAAAGAAAAATGAATGAGGCCGTCGATATTTTGGAGGAATGGCTGAAGAAACAGAATCATTTCCTGAAGAAAGATATTCCAAGAATATACTTGGAGACGAATATAATAGGAAGCAAAGGTTCTATCGAGAGAACTAAATCAAAACTGGACAAGTTGTTCGCTCTCCGGACTGTTCTACGGAAGTTTTATGAATTCAATGATATCAGAAATGAGTTTAAAAACCTGTTCTCGTGTACAAGCATGCTAGCTATGCCTAAATTGACGAAAGAAAACAATAGACTTTTTGTAGTTAAAGTGATTGGAAACGAGTTCTTTGATAAAGTTCCATATTTAGATTATTACAAAATTATTATTGCGTATGTAGAATACTTCAGGATGTATGATTACTCGGACGGATACAGAGCAGTTATTGACTACAGGGATATCAACATTATGAGTATAATAGCTCGCGTCAACCTGGTAGAGTTAAAAGAGTTTTCCACGCTTATTTTATCCGGCTACGGTGCGAGGCTCAAAGGAATCCATATAATAACTTCATCGAAGGCAGTCGAGAATTTGCTGAGAATAATAACACCGTTATTCAATCCAAAATTGGTGGCCAGATTCAGAGTTCACGGAACGTTGGAGTCATTACACGAAGTAATATCTCAGGATAGGTTACCAAAAGAGCTGGGAGGGACTGGACGATCGCTTAAAGAAATACAAGATGACTGGATCGACCTGTTGAGCTCGAAGGAGACCTCAGAATTCTTCCGCGAGATAAACAAAGCACAGGTCAACGAGGCGTATAGAATGGATGATTTGAAGAATGAAGAAATGGGATTGCCGGGAACTTTCAGAACATTAAGTGTTGATTGA
Protein
MYLTLKQKLLEVPEGTAQQIQMLYGYDERKMNEAVDILEEWLKKQNHFLKKDIPRIYLETNIIGSKGSIERTKSKLDKLFALRTVLRKFYEFNDIRNEFKNLFSCTSMLAMPKLTKENNRLFVVKVIGNEFFDKVPYLDYYKIIIAYVEYFRMYDYSDGYRAVIDYRDINIMSIIARVNLVELKEFSTLILSGYGARLKGIHIITSSKAVENLLRIITPLFNPKLVARFRVHGTLESLHEVISQDRLPKELGGTGRSLKEIQDDWIDLLSSKETSEFFREINKAQVNEAYRMDDLKNEEMGLPGTFRTLSVD
Summary
Uniprot
H9JKR5
H9JKR6
A0A0S2Z358
A0A194PZV7
A0A0S2Z340
A0A2H4RMP8
+ More
A0A2A4J6B5 A0A194PZV1 A0A2W1BF53 A0A0S2Z393 A0A0S2Z354 A0A194R247 A0A194R1M2 A0A2W1BI70 A0A194Q0M6 A0A2H1VU69 A0A212FN73 A0A2A4JNW0 A0A2H1WD56 A0A2H4RMR2 A0A0S2Z343 A0A2A4IUI0 A0A2A4JNK7 A0A2H4RMR8 A0A2H4RMT4 A0A2H4RMU5 A0A2H4RMT2 A0A2W1BCD2 A0A2H1W264 A0A3S2L7M8 A0A2A4J4G2 A0A2W1BJJ9 A0A2H4RMU0 A0A2H4RMS3 A0A2H4RMR5 A0A3S2NSP3 A0A2H1UZW6 A0A2H1VYX3 A0A2H4RMT7 A0A2A4J610 A0A2H1V3H4 A0A0S2Z362 A0A2H1VXT5 A0A212FN16 A0A2H4RMS9 A0A2W1BML1 A0A1E1WBH5 A0A2A4IW24 A0A212FN55 A0A3S2LZD4 A0A194R1B9 A0A2H1W3J3 A0A3S2NCI1 A0A2A4J0W3 A0A2H1W0Z0 A0A2A4J2F6 A0A2W1BGX0 A0A2H4RMR6 A0A2H4RMS8 A0A2H1VM17 A0A2H4RMP7 A0A2A4JMX0 A0A2H4RMV1 A0A2W1BGS3 A0A0L7L3C1 A0A0N7EI05 A0A0S2Z2V1 A0A1E1VYT1 A0A2H1WHW8 H9JTM3 A0A2H1WDV2 A0A3S2PCJ5 A0A212FN46 V5I956 A0A194R2X0 A0A194Q621 A0A2H4RMS7 V5G488 A0A2J7Q1W1 A0A3S2L872 A0A1Y1MVW2 A0A2H1V2T7 A0A2A4JLK9 A0A1J1ICM2 A0A067RJF7 E2BJ28 A0A3L8E2J1 B0WR71 Q1HQ95 A0A194Q6G5 H9JU04 A0A212FMF6 A0A2H1WHQ3
A0A2A4J6B5 A0A194PZV1 A0A2W1BF53 A0A0S2Z393 A0A0S2Z354 A0A194R247 A0A194R1M2 A0A2W1BI70 A0A194Q0M6 A0A2H1VU69 A0A212FN73 A0A2A4JNW0 A0A2H1WD56 A0A2H4RMR2 A0A0S2Z343 A0A2A4IUI0 A0A2A4JNK7 A0A2H4RMR8 A0A2H4RMT4 A0A2H4RMU5 A0A2H4RMT2 A0A2W1BCD2 A0A2H1W264 A0A3S2L7M8 A0A2A4J4G2 A0A2W1BJJ9 A0A2H4RMU0 A0A2H4RMS3 A0A2H4RMR5 A0A3S2NSP3 A0A2H1UZW6 A0A2H1VYX3 A0A2H4RMT7 A0A2A4J610 A0A2H1V3H4 A0A0S2Z362 A0A2H1VXT5 A0A212FN16 A0A2H4RMS9 A0A2W1BML1 A0A1E1WBH5 A0A2A4IW24 A0A212FN55 A0A3S2LZD4 A0A194R1B9 A0A2H1W3J3 A0A3S2NCI1 A0A2A4J0W3 A0A2H1W0Z0 A0A2A4J2F6 A0A2W1BGX0 A0A2H4RMR6 A0A2H4RMS8 A0A2H1VM17 A0A2H4RMP7 A0A2A4JMX0 A0A2H4RMV1 A0A2W1BGS3 A0A0L7L3C1 A0A0N7EI05 A0A0S2Z2V1 A0A1E1VYT1 A0A2H1WHW8 H9JTM3 A0A2H1WDV2 A0A3S2PCJ5 A0A212FN46 V5I956 A0A194R2X0 A0A194Q621 A0A2H4RMS7 V5G488 A0A2J7Q1W1 A0A3S2L872 A0A1Y1MVW2 A0A2H1V2T7 A0A2A4JLK9 A0A1J1ICM2 A0A067RJF7 E2BJ28 A0A3L8E2J1 B0WR71 Q1HQ95 A0A194Q6G5 H9JU04 A0A212FMF6 A0A2H1WHQ3
Pubmed
EMBL
BABH01011652
BABH01011650
BABH01011651
KT943545
ALQ33303.1
KQ459582
+ More
KPI98856.1 KT943544 ALQ33302.1 MG434605 ATY51911.1 NWSH01003092 PCG66963.1 KPI98857.1 KZ150143 PZC72921.1 KT943562 ALQ33320.1 KT943558 ALQ33316.1 KQ460878 KPJ11589.1 KPJ11587.1 PZC72917.1 KPI98858.1 ODYU01004475 SOQ44379.1 AGBW02007648 OWR55140.1 NWSH01000951 PCG73398.1 ODYU01007545 SOQ50404.1 MG434625 ATY51931.1 KT943542 ALQ33300.1 NWSH01006612 PCG63329.1 PCG73399.1 MG434628 ATY51934.1 MG434630 ATY51936.1 MG434650 ATY51956.1 MG434631 ATY51937.1 PZC72922.1 ODYU01005865 SOQ47185.1 RSAL01000100 RVE47537.1 PCG66965.1 PZC72920.1 MG434648 ATY51954.1 MG434627 ATY51933.1 MG434624 ATY51930.1 RVE47536.1 ODYU01000041 SOQ34143.1 ODYU01005307 SOQ46031.1 MG434643 ATY51949.1 PCG66964.1 ODYU01000500 SOQ35381.1 KT943537 ALQ33295.1 ODYU01005115 SOQ45661.1 OWR55142.1 MG434629 ATY51935.1 PZC72923.1 GDQN01006712 GDQN01000539 JAT84342.1 JAT90515.1 PCG63330.1 OWR55139.1 RVE47535.1 KPJ11588.1 ODYU01006075 SOQ47608.1 RVE47534.1 NWSH01003956 PCG65621.1 ODYU01005386 SOQ46174.1 PCG65620.1 PZC72924.1 MG434626 ATY51932.1 MG434632 ATY51938.1 ODYU01003303 SOQ41879.1 MG434603 ATY51909.1 PCG73397.1 MG434656 ATY51962.1 PZC72915.1 JTDY01003302 KOB69774.1 KT781079 MG434657 ALF12134.1 ATY51963.1 KT943555 ALQ33313.1 GDQN01011167 JAT79887.1 ODYU01008754 SOQ52627.1 BABH01003542 ODYU01008002 SOQ51248.1 RVE47533.1 OWR55143.1 GALX01003520 JAB64946.1 KPJ11590.1 KPI98855.1 MG434633 ATY51939.1 GALX01003616 JAB64850.1 NEVH01019379 PNF22549.1 RSAL01000093 RVE47936.1 GEZM01023906 GEZM01023905 GEZM01023904 GEZM01023902 GEZM01023901 JAV88146.1 ODYU01000399 SOQ35119.1 NWSH01001060 PCG72861.1 CVRI01000047 CRK97300.1 KK852648 KDR19512.1 GL448558 EFN84178.1 QOIP01000001 RLU26595.1 DS232052 EDS33245.1 DQ443157 ABF51246.1 KPI98995.1 BABH01003855 BABH01003856 AGBW02007651 OWR54932.1 ODYU01008739 SOQ52601.1
KPI98856.1 KT943544 ALQ33302.1 MG434605 ATY51911.1 NWSH01003092 PCG66963.1 KPI98857.1 KZ150143 PZC72921.1 KT943562 ALQ33320.1 KT943558 ALQ33316.1 KQ460878 KPJ11589.1 KPJ11587.1 PZC72917.1 KPI98858.1 ODYU01004475 SOQ44379.1 AGBW02007648 OWR55140.1 NWSH01000951 PCG73398.1 ODYU01007545 SOQ50404.1 MG434625 ATY51931.1 KT943542 ALQ33300.1 NWSH01006612 PCG63329.1 PCG73399.1 MG434628 ATY51934.1 MG434630 ATY51936.1 MG434650 ATY51956.1 MG434631 ATY51937.1 PZC72922.1 ODYU01005865 SOQ47185.1 RSAL01000100 RVE47537.1 PCG66965.1 PZC72920.1 MG434648 ATY51954.1 MG434627 ATY51933.1 MG434624 ATY51930.1 RVE47536.1 ODYU01000041 SOQ34143.1 ODYU01005307 SOQ46031.1 MG434643 ATY51949.1 PCG66964.1 ODYU01000500 SOQ35381.1 KT943537 ALQ33295.1 ODYU01005115 SOQ45661.1 OWR55142.1 MG434629 ATY51935.1 PZC72923.1 GDQN01006712 GDQN01000539 JAT84342.1 JAT90515.1 PCG63330.1 OWR55139.1 RVE47535.1 KPJ11588.1 ODYU01006075 SOQ47608.1 RVE47534.1 NWSH01003956 PCG65621.1 ODYU01005386 SOQ46174.1 PCG65620.1 PZC72924.1 MG434626 ATY51932.1 MG434632 ATY51938.1 ODYU01003303 SOQ41879.1 MG434603 ATY51909.1 PCG73397.1 MG434656 ATY51962.1 PZC72915.1 JTDY01003302 KOB69774.1 KT781079 MG434657 ALF12134.1 ATY51963.1 KT943555 ALQ33313.1 GDQN01011167 JAT79887.1 ODYU01008754 SOQ52627.1 BABH01003542 ODYU01008002 SOQ51248.1 RVE47533.1 OWR55143.1 GALX01003520 JAB64946.1 KPJ11590.1 KPI98855.1 MG434633 ATY51939.1 GALX01003616 JAB64850.1 NEVH01019379 PNF22549.1 RSAL01000093 RVE47936.1 GEZM01023906 GEZM01023905 GEZM01023904 GEZM01023902 GEZM01023901 JAV88146.1 ODYU01000399 SOQ35119.1 NWSH01001060 PCG72861.1 CVRI01000047 CRK97300.1 KK852648 KDR19512.1 GL448558 EFN84178.1 QOIP01000001 RLU26595.1 DS232052 EDS33245.1 DQ443157 ABF51246.1 KPI98995.1 BABH01003855 BABH01003856 AGBW02007651 OWR54932.1 ODYU01008739 SOQ52601.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JKR5
H9JKR6
A0A0S2Z358
A0A194PZV7
A0A0S2Z340
A0A2H4RMP8
+ More
A0A2A4J6B5 A0A194PZV1 A0A2W1BF53 A0A0S2Z393 A0A0S2Z354 A0A194R247 A0A194R1M2 A0A2W1BI70 A0A194Q0M6 A0A2H1VU69 A0A212FN73 A0A2A4JNW0 A0A2H1WD56 A0A2H4RMR2 A0A0S2Z343 A0A2A4IUI0 A0A2A4JNK7 A0A2H4RMR8 A0A2H4RMT4 A0A2H4RMU5 A0A2H4RMT2 A0A2W1BCD2 A0A2H1W264 A0A3S2L7M8 A0A2A4J4G2 A0A2W1BJJ9 A0A2H4RMU0 A0A2H4RMS3 A0A2H4RMR5 A0A3S2NSP3 A0A2H1UZW6 A0A2H1VYX3 A0A2H4RMT7 A0A2A4J610 A0A2H1V3H4 A0A0S2Z362 A0A2H1VXT5 A0A212FN16 A0A2H4RMS9 A0A2W1BML1 A0A1E1WBH5 A0A2A4IW24 A0A212FN55 A0A3S2LZD4 A0A194R1B9 A0A2H1W3J3 A0A3S2NCI1 A0A2A4J0W3 A0A2H1W0Z0 A0A2A4J2F6 A0A2W1BGX0 A0A2H4RMR6 A0A2H4RMS8 A0A2H1VM17 A0A2H4RMP7 A0A2A4JMX0 A0A2H4RMV1 A0A2W1BGS3 A0A0L7L3C1 A0A0N7EI05 A0A0S2Z2V1 A0A1E1VYT1 A0A2H1WHW8 H9JTM3 A0A2H1WDV2 A0A3S2PCJ5 A0A212FN46 V5I956 A0A194R2X0 A0A194Q621 A0A2H4RMS7 V5G488 A0A2J7Q1W1 A0A3S2L872 A0A1Y1MVW2 A0A2H1V2T7 A0A2A4JLK9 A0A1J1ICM2 A0A067RJF7 E2BJ28 A0A3L8E2J1 B0WR71 Q1HQ95 A0A194Q6G5 H9JU04 A0A212FMF6 A0A2H1WHQ3
A0A2A4J6B5 A0A194PZV1 A0A2W1BF53 A0A0S2Z393 A0A0S2Z354 A0A194R247 A0A194R1M2 A0A2W1BI70 A0A194Q0M6 A0A2H1VU69 A0A212FN73 A0A2A4JNW0 A0A2H1WD56 A0A2H4RMR2 A0A0S2Z343 A0A2A4IUI0 A0A2A4JNK7 A0A2H4RMR8 A0A2H4RMT4 A0A2H4RMU5 A0A2H4RMT2 A0A2W1BCD2 A0A2H1W264 A0A3S2L7M8 A0A2A4J4G2 A0A2W1BJJ9 A0A2H4RMU0 A0A2H4RMS3 A0A2H4RMR5 A0A3S2NSP3 A0A2H1UZW6 A0A2H1VYX3 A0A2H4RMT7 A0A2A4J610 A0A2H1V3H4 A0A0S2Z362 A0A2H1VXT5 A0A212FN16 A0A2H4RMS9 A0A2W1BML1 A0A1E1WBH5 A0A2A4IW24 A0A212FN55 A0A3S2LZD4 A0A194R1B9 A0A2H1W3J3 A0A3S2NCI1 A0A2A4J0W3 A0A2H1W0Z0 A0A2A4J2F6 A0A2W1BGX0 A0A2H4RMR6 A0A2H4RMS8 A0A2H1VM17 A0A2H4RMP7 A0A2A4JMX0 A0A2H4RMV1 A0A2W1BGS3 A0A0L7L3C1 A0A0N7EI05 A0A0S2Z2V1 A0A1E1VYT1 A0A2H1WHW8 H9JTM3 A0A2H1WDV2 A0A3S2PCJ5 A0A212FN46 V5I956 A0A194R2X0 A0A194Q621 A0A2H4RMS7 V5G488 A0A2J7Q1W1 A0A3S2L872 A0A1Y1MVW2 A0A2H1V2T7 A0A2A4JLK9 A0A1J1ICM2 A0A067RJF7 E2BJ28 A0A3L8E2J1 B0WR71 Q1HQ95 A0A194Q6G5 H9JU04 A0A212FMF6 A0A2H1WHQ3
PDB
4CIZ
E-value=0.0030013,
Score=95
Ontologies
GO
Topology
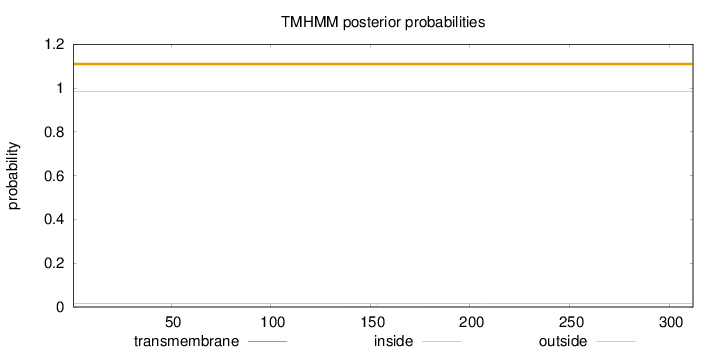
Length:
312
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01122
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01447
outside
1 - 312
Population Genetic Test Statistics
Pi
225.128852
Theta
170.303057
Tajima's D
1.345687
CLR
0.000054
CSRT
0.752412379381031
Interpretation
Uncertain
