Gene
KWMTBOMO04010
Pre Gene Modal
BGIBMGA010118
Annotation
PREDICTED:_uncharacterized_protein_LOC106131606_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.776
Sequence
CDS
ATGGTTTTAATCATAAAGTCAGTAGTACGCTCGTATCGGTCTACTATCCATATCATTAGATCGTACTGCGACAATAAATCAATTTATCAAAAAAATGAAGAGGGTCCACAACCCCGAATAATTCACGGCCAAGTCTATCCTGATTGGAGGAAACCATGGATTGAGAGGCAAGGAGAATGGAGAAGCAAATTATCAGTTTTTGTAGAAAAAAATCCTAGTCCAGACGTTTTATACGCTATGTCACAAATACCAAATTTAAATTTTAAAATAGTCAGAGAATGGTGGGCTCACATGAAAAACATTCAAGAAGTGGAAAATCAGAAGTATCTCCATGAAAGAGTTGCTGCTTTGGGTTTCAATTTGGCTGCATTACATTTCTTCACATACAGACATGCTGCTGTCAGATTAAAAGATTCAAGCGACTGGTTGAAAGGTGATATACTATCTCTGAAACTACCAGATCATTATACTGAGGGTTATTATGTTGAAGCTATTGACTGCTCAAATTTCCATCACGGTGGCATTCGCTATGAAGGCCTAGAAAATCTAAAAGGTCTCAATTTATTGAAATGGTTATCTTTAAGAAATAACAAACACGTTGATGTTTGGTGCTTAGATAGAGTAGCGGGATTGTCAGGCCAATCCTTAGAATATCTTGACATTAGAGGTTGTAAAATTTGCATTGGATGTATTAATGCATTAGCGAGAATGGAGGTTTTGAAATACTTACTTGTCTCTGATCCTGGTGATGACATGGCTCTTCAAGCAGGACTGTCAGTGTTAGAAGAAAACAATCCAAATTTGTTCATAAAAGCTGACTGA
Protein
MVLIIKSVVRSYRSTIHIIRSYCDNKSIYQKNEEGPQPRIIHGQVYPDWRKPWIERQGEWRSKLSVFVEKNPSPDVLYAMSQIPNLNFKIVREWWAHMKNIQEVENQKYLHERVAALGFNLAALHFFTYRHAAVRLKDSSDWLKGDILSLKLPDHYTEGYYVEAIDCSNFHHGGIRYEGLENLKGLNLLKWLSLRNNKHVDVWCLDRVAGLSGQSLEYLDIRGCKICIGCINALARMEVLKYLLVSDPGDDMALQAGLSVLEENNPNLFIKAD
Summary
Uniprot
A0A3S2NEX1
A0A2W1BKM2
A0A194RMB9
A0A2A4JUU8
A0A0L7L4D6
S4PAC8
+ More
I4DL94 A0A194PLQ7 A0A2H1WUI2 A0A2J7QT77 A0A034WR05 W8B2F4 A0A2P8YVI4 Q17DN6 A0A182N3T3 A0A182M4C0 A0A0K8UBP0 A0A182PHB0 A0A182QT40 A0A182YBI0 A0A084WLY3 Q5TX70 A0A1L8E048 B0W815 A0A182WQN5 B4QJ64 B4IAF0 A0A182RUJ7 A0A182J8B4 A0A182V4H4 A0A182S685 B4PER1 B4IUI0 A0A1W4VMX3 A0A182HW18 A0A067R1Z7 B3M9S6 A0A182WUG0 A0A182JS76 Q9VPC1 Q29DA5 W5J972 B3NIR1 A0A139WL00 A0A1B0GHF9 B4N4J8 Q8SYK3 A0A0M5J8K4 A0A2M3ZJF5 A0A2M4BWP7 A0A1Q3F849 A0A1Y1LAC4 B4KYH5 A0A3B0JSZ0 A0A2M4BWH6 A0A2M4BWI7 B4J3N5 A0A0L0C925 A0A1B0D3R3 A0A023ELZ4 B4LG21 A0A1W4WU24 J3JY80 A0A1I8M9P7 A0A1B6JHN1 A0A1J1IQY1 A0A1B6ERZ6 B4H7R8 A0A1I8PHR3 A0A182FUW6 A0A0N8AID6 A0A162RKB3 A0A0P6HZ58 A0A0P5E4A2 A0A0P5YNJ2 A0A0V0G6K3 A0A0P6C6G6 A0A1B6DA94 A0A151IEL8 A0A0J7KX94 A0A336M527 J9KU35 A0A2S2P6W5 A0A2H8TXN2 A0A1B0FA42 E9IRE4 A0A158NKM5
I4DL94 A0A194PLQ7 A0A2H1WUI2 A0A2J7QT77 A0A034WR05 W8B2F4 A0A2P8YVI4 Q17DN6 A0A182N3T3 A0A182M4C0 A0A0K8UBP0 A0A182PHB0 A0A182QT40 A0A182YBI0 A0A084WLY3 Q5TX70 A0A1L8E048 B0W815 A0A182WQN5 B4QJ64 B4IAF0 A0A182RUJ7 A0A182J8B4 A0A182V4H4 A0A182S685 B4PER1 B4IUI0 A0A1W4VMX3 A0A182HW18 A0A067R1Z7 B3M9S6 A0A182WUG0 A0A182JS76 Q9VPC1 Q29DA5 W5J972 B3NIR1 A0A139WL00 A0A1B0GHF9 B4N4J8 Q8SYK3 A0A0M5J8K4 A0A2M3ZJF5 A0A2M4BWP7 A0A1Q3F849 A0A1Y1LAC4 B4KYH5 A0A3B0JSZ0 A0A2M4BWH6 A0A2M4BWI7 B4J3N5 A0A0L0C925 A0A1B0D3R3 A0A023ELZ4 B4LG21 A0A1W4WU24 J3JY80 A0A1I8M9P7 A0A1B6JHN1 A0A1J1IQY1 A0A1B6ERZ6 B4H7R8 A0A1I8PHR3 A0A182FUW6 A0A0N8AID6 A0A162RKB3 A0A0P6HZ58 A0A0P5E4A2 A0A0P5YNJ2 A0A0V0G6K3 A0A0P6C6G6 A0A1B6DA94 A0A151IEL8 A0A0J7KX94 A0A336M527 J9KU35 A0A2S2P6W5 A0A2H8TXN2 A0A1B0FA42 E9IRE4 A0A158NKM5
Pubmed
28756777
26354079
26227816
23622113
22651552
25348373
+ More
24495485 29403074 17510324 25244985 24438588 12364791 17994087 22936249 17550304 24845553 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20920257 23761445 18362917 19820115 28004739 26108605 24945155 22516182 25315136 21282665 21347285
24495485 29403074 17510324 25244985 24438588 12364791 17994087 22936249 17550304 24845553 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20920257 23761445 18362917 19820115 28004739 26108605 24945155 22516182 25315136 21282665 21347285
EMBL
RSAL01000067
RVE49334.1
KZ150064
PZC74184.1
KQ459995
KPJ18569.1
+ More
NWSH01000620 PCG75263.1 JTDY01002986 KOB70363.1 GAIX01003314 JAA89246.1 AK402062 BAM18684.1 KQ459600 KPI94262.1 ODYU01011158 SOQ56729.1 NEVH01011202 PNF31773.1 GAKP01002774 JAC56178.1 GAMC01013838 JAB92717.1 PYGN01000337 PSN48258.1 CH477292 EAT44539.1 AXCM01001195 GDHF01028323 JAI23991.1 AXCN02001098 ATLV01024321 KE525351 KFB51227.1 AAAB01008804 EAL41993.3 GFDF01002023 JAV12061.1 DS231856 EDS38477.1 CM000363 CM002912 EDX11281.1 KMZ00851.1 CH480826 EDW44263.1 CM000159 EDW95135.1 CH891918 EDX00044.1 APCN01001457 KK852771 KDR16882.1 CH902618 EDV40117.1 KPU78377.1 AE014296 BT044316 AAF51634.1 ACH92381.1 CH379070 EAL30509.2 ADMH02001912 ETN60536.1 CH954178 EDV52557.1 KQ971321 KYB28622.1 AJWK01004553 CH964095 EDW79072.1 AY071495 AAL49117.1 CP012525 ALC44786.1 GGFM01007913 MBW28664.1 GGFJ01008293 MBW57434.1 GFDL01011362 JAV23683.1 GEZM01065183 JAV68536.1 CH933809 EDW18786.2 OUUW01000002 SPP76819.1 GGFJ01008294 MBW57435.1 GGFJ01008295 MBW57436.1 CH916366 EDV97266.1 JRES01000836 KNC27884.1 AJVK01023648 GAPW01003296 JAC10302.1 CH940647 EDW70420.1 BT128204 AEE63165.1 GECU01008900 JAS98806.1 CVRI01000058 CRL02552.1 GECZ01029089 JAS40680.1 CH479219 EDW34708.1 GDIP01144449 JAJ78953.1 LRGB01000146 KZS20584.1 GDIQ01019825 JAN74912.1 GDIP01165280 JAJ58122.1 GDIP01056668 JAM47047.1 GECL01002454 JAP03670.1 GDIP01012725 JAM90990.1 GEDC01014793 JAS22505.1 KQ977862 KYM99267.1 LBMM01002347 KMQ94946.1 UFQT01000174 SSX21128.1 ABLF02027507 GGMR01012545 MBY25164.1 GFXV01007201 MBW19006.1 CCAG010023047 GL765107 EFZ16892.1 ADTU01018888
NWSH01000620 PCG75263.1 JTDY01002986 KOB70363.1 GAIX01003314 JAA89246.1 AK402062 BAM18684.1 KQ459600 KPI94262.1 ODYU01011158 SOQ56729.1 NEVH01011202 PNF31773.1 GAKP01002774 JAC56178.1 GAMC01013838 JAB92717.1 PYGN01000337 PSN48258.1 CH477292 EAT44539.1 AXCM01001195 GDHF01028323 JAI23991.1 AXCN02001098 ATLV01024321 KE525351 KFB51227.1 AAAB01008804 EAL41993.3 GFDF01002023 JAV12061.1 DS231856 EDS38477.1 CM000363 CM002912 EDX11281.1 KMZ00851.1 CH480826 EDW44263.1 CM000159 EDW95135.1 CH891918 EDX00044.1 APCN01001457 KK852771 KDR16882.1 CH902618 EDV40117.1 KPU78377.1 AE014296 BT044316 AAF51634.1 ACH92381.1 CH379070 EAL30509.2 ADMH02001912 ETN60536.1 CH954178 EDV52557.1 KQ971321 KYB28622.1 AJWK01004553 CH964095 EDW79072.1 AY071495 AAL49117.1 CP012525 ALC44786.1 GGFM01007913 MBW28664.1 GGFJ01008293 MBW57434.1 GFDL01011362 JAV23683.1 GEZM01065183 JAV68536.1 CH933809 EDW18786.2 OUUW01000002 SPP76819.1 GGFJ01008294 MBW57435.1 GGFJ01008295 MBW57436.1 CH916366 EDV97266.1 JRES01000836 KNC27884.1 AJVK01023648 GAPW01003296 JAC10302.1 CH940647 EDW70420.1 BT128204 AEE63165.1 GECU01008900 JAS98806.1 CVRI01000058 CRL02552.1 GECZ01029089 JAS40680.1 CH479219 EDW34708.1 GDIP01144449 JAJ78953.1 LRGB01000146 KZS20584.1 GDIQ01019825 JAN74912.1 GDIP01165280 JAJ58122.1 GDIP01056668 JAM47047.1 GECL01002454 JAP03670.1 GDIP01012725 JAM90990.1 GEDC01014793 JAS22505.1 KQ977862 KYM99267.1 LBMM01002347 KMQ94946.1 UFQT01000174 SSX21128.1 ABLF02027507 GGMR01012545 MBY25164.1 GFXV01007201 MBW19006.1 CCAG010023047 GL765107 EFZ16892.1 ADTU01018888
Proteomes
UP000283053
UP000053240
UP000218220
UP000037510
UP000053268
UP000235965
+ More
UP000245037 UP000008820 UP000075884 UP000075883 UP000075885 UP000075886 UP000076408 UP000030765 UP000007062 UP000002320 UP000075920 UP000000304 UP000001292 UP000075900 UP000075880 UP000075903 UP000075901 UP000002282 UP000192221 UP000075840 UP000027135 UP000007801 UP000076407 UP000075881 UP000000803 UP000001819 UP000000673 UP000008711 UP000007266 UP000092461 UP000007798 UP000092553 UP000009192 UP000268350 UP000001070 UP000037069 UP000092462 UP000008792 UP000192223 UP000095301 UP000183832 UP000008744 UP000095300 UP000069272 UP000076858 UP000078542 UP000036403 UP000007819 UP000092444 UP000005205
UP000245037 UP000008820 UP000075884 UP000075883 UP000075885 UP000075886 UP000076408 UP000030765 UP000007062 UP000002320 UP000075920 UP000000304 UP000001292 UP000075900 UP000075880 UP000075903 UP000075901 UP000002282 UP000192221 UP000075840 UP000027135 UP000007801 UP000076407 UP000075881 UP000000803 UP000001819 UP000000673 UP000008711 UP000007266 UP000092461 UP000007798 UP000092553 UP000009192 UP000268350 UP000001070 UP000037069 UP000092462 UP000008792 UP000192223 UP000095301 UP000183832 UP000008744 UP000095300 UP000069272 UP000076858 UP000078542 UP000036403 UP000007819 UP000092444 UP000005205
Interpro
Gene 3D
ProteinModelPortal
A0A3S2NEX1
A0A2W1BKM2
A0A194RMB9
A0A2A4JUU8
A0A0L7L4D6
S4PAC8
+ More
I4DL94 A0A194PLQ7 A0A2H1WUI2 A0A2J7QT77 A0A034WR05 W8B2F4 A0A2P8YVI4 Q17DN6 A0A182N3T3 A0A182M4C0 A0A0K8UBP0 A0A182PHB0 A0A182QT40 A0A182YBI0 A0A084WLY3 Q5TX70 A0A1L8E048 B0W815 A0A182WQN5 B4QJ64 B4IAF0 A0A182RUJ7 A0A182J8B4 A0A182V4H4 A0A182S685 B4PER1 B4IUI0 A0A1W4VMX3 A0A182HW18 A0A067R1Z7 B3M9S6 A0A182WUG0 A0A182JS76 Q9VPC1 Q29DA5 W5J972 B3NIR1 A0A139WL00 A0A1B0GHF9 B4N4J8 Q8SYK3 A0A0M5J8K4 A0A2M3ZJF5 A0A2M4BWP7 A0A1Q3F849 A0A1Y1LAC4 B4KYH5 A0A3B0JSZ0 A0A2M4BWH6 A0A2M4BWI7 B4J3N5 A0A0L0C925 A0A1B0D3R3 A0A023ELZ4 B4LG21 A0A1W4WU24 J3JY80 A0A1I8M9P7 A0A1B6JHN1 A0A1J1IQY1 A0A1B6ERZ6 B4H7R8 A0A1I8PHR3 A0A182FUW6 A0A0N8AID6 A0A162RKB3 A0A0P6HZ58 A0A0P5E4A2 A0A0P5YNJ2 A0A0V0G6K3 A0A0P6C6G6 A0A1B6DA94 A0A151IEL8 A0A0J7KX94 A0A336M527 J9KU35 A0A2S2P6W5 A0A2H8TXN2 A0A1B0FA42 E9IRE4 A0A158NKM5
I4DL94 A0A194PLQ7 A0A2H1WUI2 A0A2J7QT77 A0A034WR05 W8B2F4 A0A2P8YVI4 Q17DN6 A0A182N3T3 A0A182M4C0 A0A0K8UBP0 A0A182PHB0 A0A182QT40 A0A182YBI0 A0A084WLY3 Q5TX70 A0A1L8E048 B0W815 A0A182WQN5 B4QJ64 B4IAF0 A0A182RUJ7 A0A182J8B4 A0A182V4H4 A0A182S685 B4PER1 B4IUI0 A0A1W4VMX3 A0A182HW18 A0A067R1Z7 B3M9S6 A0A182WUG0 A0A182JS76 Q9VPC1 Q29DA5 W5J972 B3NIR1 A0A139WL00 A0A1B0GHF9 B4N4J8 Q8SYK3 A0A0M5J8K4 A0A2M3ZJF5 A0A2M4BWP7 A0A1Q3F849 A0A1Y1LAC4 B4KYH5 A0A3B0JSZ0 A0A2M4BWH6 A0A2M4BWI7 B4J3N5 A0A0L0C925 A0A1B0D3R3 A0A023ELZ4 B4LG21 A0A1W4WU24 J3JY80 A0A1I8M9P7 A0A1B6JHN1 A0A1J1IQY1 A0A1B6ERZ6 B4H7R8 A0A1I8PHR3 A0A182FUW6 A0A0N8AID6 A0A162RKB3 A0A0P6HZ58 A0A0P5E4A2 A0A0P5YNJ2 A0A0V0G6K3 A0A0P6C6G6 A0A1B6DA94 A0A151IEL8 A0A0J7KX94 A0A336M527 J9KU35 A0A2S2P6W5 A0A2H8TXN2 A0A1B0FA42 E9IRE4 A0A158NKM5
Ontologies
PANTHER
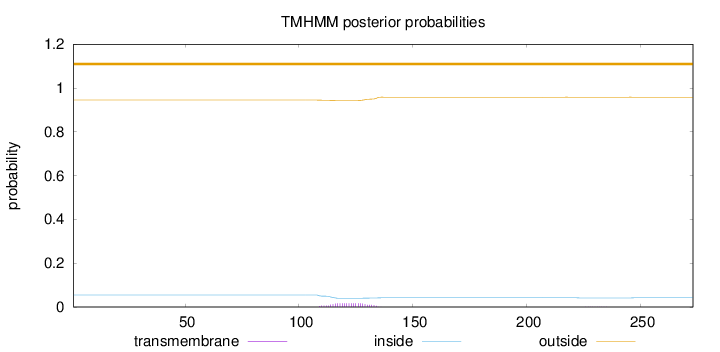
Topology
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32967
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05490
outside
1 - 273
Population Genetic Test Statistics
Pi
479.253303
Theta
236.187011
Tajima's D
3.222044
CLR
0
CSRT
0.986850657467127
Interpretation
Uncertain
