Pre Gene Modal
BGIBMGA010263
Annotation
PREDICTED:_putative_transcription_factor_capicua_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.581
Sequence
CDS
ATGGCCAATCCGACTAATCTCGGCAACGCCAGCAGCGGCAACGCTACTCAGTCGACGGTCCATCAAACGCCAACTCAAAACCCAGTGAGGAATCTGCCCAAGAAACGCAAATTTGACCCTTCCTCGCTAGAGGAGATGCAGCAGACCTGTCCCAATAATAACACGATCAACACTTCGACGGCGTCGGTACTCGACTTCCACAACTATCAACGGCCAGTGCTCCAACCATCGCCAATTGTACGGCAATCTCCACCCCAAGACGTGAAGCCGTTTTTCATCCAATACCCAAACATTGATTTGTCCGAATGGCGTGACCACCGAGTTTTGGCCAAGCAACGGGGCTTTTATGTACCCGGAGCGATAAGACATGCCGAAGGTTGTAAAGTGACAGTAGAGTTAGACGGTTATGAAAACGAACCTGTCGAATACAGTGACGTGTTTGGTATGAACAAACTGGATGTTATCGGCGATGCCAGCCCGCAGATGAGCCATTTGCTGATGGGTCTGCCGTGCGTCGTGCGTACCTCTGACCCCAATCGAGAAAACGTTCAAAACGTATTCGTCGAAGGTTTCATCTTAGAGGTGTTAAATTCGCCTATAAGGATACGCGTTGAGGTGAGAGATGGCGACCTATGCAAGCAGGCGGTGGAAGTGAAGCGGGCGGATATCCGGCTGATGCAGCCACCCTGGGCTGATGAACTGGAAGACGCCGGCTCCCACGTGTCCTCGTCACTGCACGCGCATCCAATGCGAGTACACCATCCGCCGCATCAGATCGGCGGCGACCACTTCTACACTTCGTCTCCGATGCCCGGGGCCGGTTCGGGCGTGGTCACGGTCGGGGCGCTTTCGAACGGTTCGATGGACGAGCTCAGGAAGCGAACCTTCGACGACTACCCGGAGAGCGATGACGAATTGCGCCGCGAGGACATCATGTTCTCAACTGACGTATCGCACATGGGTGAGTGA
Protein
MANPTNLGNASSGNATQSTVHQTPTQNPVRNLPKKRKFDPSSLEEMQQTCPNNNTINTSTASVLDFHNYQRPVLQPSPIVRQSPPQDVKPFFIQYPNIDLSEWRDHRVLAKQRGFYVPGAIRHAEGCKVTVELDGYENEPVEYSDVFGMNKLDVIGDASPQMSHLLMGLPCVVRTSDPNRENVQNVFVEGFILEVLNSPIRIRVEVRDGDLCKQAVEVKRADIRLMQPPWADELEDAGSHVSSSLHAHPMRVHHPPHQIGGDHFYTSSPMPGAGSGVVTVGALSNGSMDELRKRTFDDYPESDDELRREDIMFSTDVSHMGE
Summary
Uniprot
H9JL62
A0A2H1VJH6
A0A2W1BGI5
A0A2A4JTC4
A0A2A4JV37
A0A2A4JV20
+ More
A0A2A4JTW6 A0A0L7L511 A0A194RRA7 A0A194PLE9 A0A212EUE2 A0A1Y1MDL5 E0VC47 A0A067R1P1 V5GBY2 A0A1B6KEX8 A0A2J7QT55 A0A1B6D1A4 A0A2R7WQ87 A0A3Q0JIE1 D6W6A1 A0A023F153 A0A1L7RSY9 A0A2P8Z674 A0A2S2R6V4 A0A158NZV7 A0A195BMV8 F4WID3 A0A026WRL2 J9JWA3 A0A3L8DVD5 A0A195F2M3 A0A154P9L6 A0A2S2P1Z1 A0A151J2H8 A0A195CMQ8 E2ADM3 A0A151XBJ7 U4UNW6 A0A1B6F3P1 A0A087ZX56 A0A146LBQ3 A0A146LTC1 A0A0A9YPM5 T1HAN5 E2B6U9 A0A1B0CSA7 A0A0C9RTH3 A0A336K074 A0A232FHC4 A0A162NUD0 A0A0P5MP10 A0A0P6FNZ8 A0A0N8AB22 A0A0P5T074 A0A0P5M7R7 K7IPC2 A0A1B6HPX7 A0A0P6BQD7 A0A226DAQ7 A0A0P6I3D2 A0A087T8X8
A0A2A4JTW6 A0A0L7L511 A0A194RRA7 A0A194PLE9 A0A212EUE2 A0A1Y1MDL5 E0VC47 A0A067R1P1 V5GBY2 A0A1B6KEX8 A0A2J7QT55 A0A1B6D1A4 A0A2R7WQ87 A0A3Q0JIE1 D6W6A1 A0A023F153 A0A1L7RSY9 A0A2P8Z674 A0A2S2R6V4 A0A158NZV7 A0A195BMV8 F4WID3 A0A026WRL2 J9JWA3 A0A3L8DVD5 A0A195F2M3 A0A154P9L6 A0A2S2P1Z1 A0A151J2H8 A0A195CMQ8 E2ADM3 A0A151XBJ7 U4UNW6 A0A1B6F3P1 A0A087ZX56 A0A146LBQ3 A0A146LTC1 A0A0A9YPM5 T1HAN5 E2B6U9 A0A1B0CSA7 A0A0C9RTH3 A0A336K074 A0A232FHC4 A0A162NUD0 A0A0P5MP10 A0A0P6FNZ8 A0A0N8AB22 A0A0P5T074 A0A0P5M7R7 K7IPC2 A0A1B6HPX7 A0A0P6BQD7 A0A226DAQ7 A0A0P6I3D2 A0A087T8X8
Pubmed
EMBL
BABH01011643
BABH01011644
BABH01011645
BABH01011646
ODYU01002639
SOQ40404.1
+ More
KZ150064 PZC74182.1 NWSH01000620 PCG75261.1 PCG75262.1 PCG75260.1 PCG75259.1 JTDY01002986 KOB70364.1 KQ459995 KPJ18571.1 KQ459600 KPI94261.1 AGBW02012402 OWR45113.1 GEZM01036449 GEZM01036448 JAV82690.1 DS235047 EEB10953.1 KK852771 KDR16881.1 GALX01000801 JAB67665.1 GEBQ01030243 JAT09734.1 NEVH01011202 PNF31757.1 GEDC01017830 JAS19468.1 KK855216 PTY21491.1 KQ971306 EFA11573.1 GBBI01003961 JAC14751.1 LN623700 CEG62428.1 PYGN01000176 PSN51999.1 GGMS01015889 MBY85092.1 ADTU01005030 KQ976440 KYM86502.1 GL888175 EGI66064.1 KK107139 EZA57739.1 ABLF02026651 QOIP01000003 RLU24397.1 KQ981856 KYN34636.1 KQ434835 KZC07918.1 GGMR01010775 MBY23394.1 KQ980404 KYN16206.1 KQ977580 KYN01752.1 GL438820 EFN68385.1 KQ982330 KYQ57668.1 KB632399 ERL94807.1 GECZ01024890 JAS44879.1 GDHC01012875 JAQ05754.1 GDHC01008959 JAQ09670.1 GBHO01010556 JAG33048.1 ACPB03012381 GL446009 EFN88590.1 AJWK01025856 AJWK01025857 GBYB01012045 JAG81812.1 UFQS01000027 UFQT01000027 SSW97767.1 SSX18153.1 NNAY01000229 OXU29858.1 LRGB01000536 KZS18269.1 GDIQ01164954 JAK86771.1 GDIQ01055286 JAN39451.1 GDIP01164961 GDIP01084623 JAJ58441.1 GDIP01133016 JAL70698.1 GDIQ01162246 JAK89479.1 GECU01030968 GECU01014157 JAS76738.1 JAS93549.1 GDIP01015062 JAM88653.1 LNIX01000025 OXA42622.1 GDIQ01011099 JAN83638.1 KK114024 KFM61567.1
KZ150064 PZC74182.1 NWSH01000620 PCG75261.1 PCG75262.1 PCG75260.1 PCG75259.1 JTDY01002986 KOB70364.1 KQ459995 KPJ18571.1 KQ459600 KPI94261.1 AGBW02012402 OWR45113.1 GEZM01036449 GEZM01036448 JAV82690.1 DS235047 EEB10953.1 KK852771 KDR16881.1 GALX01000801 JAB67665.1 GEBQ01030243 JAT09734.1 NEVH01011202 PNF31757.1 GEDC01017830 JAS19468.1 KK855216 PTY21491.1 KQ971306 EFA11573.1 GBBI01003961 JAC14751.1 LN623700 CEG62428.1 PYGN01000176 PSN51999.1 GGMS01015889 MBY85092.1 ADTU01005030 KQ976440 KYM86502.1 GL888175 EGI66064.1 KK107139 EZA57739.1 ABLF02026651 QOIP01000003 RLU24397.1 KQ981856 KYN34636.1 KQ434835 KZC07918.1 GGMR01010775 MBY23394.1 KQ980404 KYN16206.1 KQ977580 KYN01752.1 GL438820 EFN68385.1 KQ982330 KYQ57668.1 KB632399 ERL94807.1 GECZ01024890 JAS44879.1 GDHC01012875 JAQ05754.1 GDHC01008959 JAQ09670.1 GBHO01010556 JAG33048.1 ACPB03012381 GL446009 EFN88590.1 AJWK01025856 AJWK01025857 GBYB01012045 JAG81812.1 UFQS01000027 UFQT01000027 SSW97767.1 SSX18153.1 NNAY01000229 OXU29858.1 LRGB01000536 KZS18269.1 GDIQ01164954 JAK86771.1 GDIQ01055286 JAN39451.1 GDIP01164961 GDIP01084623 JAJ58441.1 GDIP01133016 JAL70698.1 GDIQ01162246 JAK89479.1 GECU01030968 GECU01014157 JAS76738.1 JAS93549.1 GDIP01015062 JAM88653.1 LNIX01000025 OXA42622.1 GDIQ01011099 JAN83638.1 KK114024 KFM61567.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000009046 UP000027135 UP000235965 UP000079169 UP000007266 UP000245037 UP000005205 UP000078540 UP000007755 UP000053097 UP000007819 UP000279307 UP000078541 UP000076502 UP000078492 UP000078542 UP000000311 UP000075809 UP000030742 UP000005203 UP000015103 UP000008237 UP000092461 UP000215335 UP000076858 UP000002358 UP000198287 UP000054359
UP000009046 UP000027135 UP000235965 UP000079169 UP000007266 UP000245037 UP000005205 UP000078540 UP000007755 UP000053097 UP000007819 UP000279307 UP000078541 UP000076502 UP000078492 UP000078542 UP000000311 UP000075809 UP000030742 UP000005203 UP000015103 UP000008237 UP000092461 UP000215335 UP000076858 UP000002358 UP000198287 UP000054359
PRIDE
SUPFAM
SSF47095
SSF47095
Gene 3D
ProteinModelPortal
H9JL62
A0A2H1VJH6
A0A2W1BGI5
A0A2A4JTC4
A0A2A4JV37
A0A2A4JV20
+ More
A0A2A4JTW6 A0A0L7L511 A0A194RRA7 A0A194PLE9 A0A212EUE2 A0A1Y1MDL5 E0VC47 A0A067R1P1 V5GBY2 A0A1B6KEX8 A0A2J7QT55 A0A1B6D1A4 A0A2R7WQ87 A0A3Q0JIE1 D6W6A1 A0A023F153 A0A1L7RSY9 A0A2P8Z674 A0A2S2R6V4 A0A158NZV7 A0A195BMV8 F4WID3 A0A026WRL2 J9JWA3 A0A3L8DVD5 A0A195F2M3 A0A154P9L6 A0A2S2P1Z1 A0A151J2H8 A0A195CMQ8 E2ADM3 A0A151XBJ7 U4UNW6 A0A1B6F3P1 A0A087ZX56 A0A146LBQ3 A0A146LTC1 A0A0A9YPM5 T1HAN5 E2B6U9 A0A1B0CSA7 A0A0C9RTH3 A0A336K074 A0A232FHC4 A0A162NUD0 A0A0P5MP10 A0A0P6FNZ8 A0A0N8AB22 A0A0P5T074 A0A0P5M7R7 K7IPC2 A0A1B6HPX7 A0A0P6BQD7 A0A226DAQ7 A0A0P6I3D2 A0A087T8X8
A0A2A4JTW6 A0A0L7L511 A0A194RRA7 A0A194PLE9 A0A212EUE2 A0A1Y1MDL5 E0VC47 A0A067R1P1 V5GBY2 A0A1B6KEX8 A0A2J7QT55 A0A1B6D1A4 A0A2R7WQ87 A0A3Q0JIE1 D6W6A1 A0A023F153 A0A1L7RSY9 A0A2P8Z674 A0A2S2R6V4 A0A158NZV7 A0A195BMV8 F4WID3 A0A026WRL2 J9JWA3 A0A3L8DVD5 A0A195F2M3 A0A154P9L6 A0A2S2P1Z1 A0A151J2H8 A0A195CMQ8 E2ADM3 A0A151XBJ7 U4UNW6 A0A1B6F3P1 A0A087ZX56 A0A146LBQ3 A0A146LTC1 A0A0A9YPM5 T1HAN5 E2B6U9 A0A1B0CSA7 A0A0C9RTH3 A0A336K074 A0A232FHC4 A0A162NUD0 A0A0P5MP10 A0A0P6FNZ8 A0A0N8AB22 A0A0P5T074 A0A0P5M7R7 K7IPC2 A0A1B6HPX7 A0A0P6BQD7 A0A226DAQ7 A0A0P6I3D2 A0A087T8X8
Ontologies
KEGG
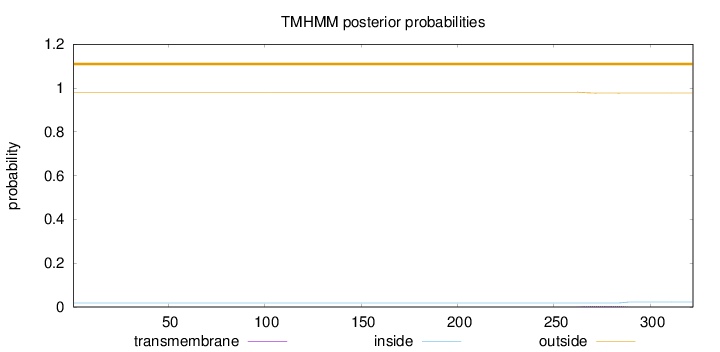
Topology
Length:
322
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09241
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01870
outside
1 - 322
Population Genetic Test Statistics
Pi
292.68451
Theta
163.868785
Tajima's D
2.670388
CLR
0.148858
CSRT
0.960801959902005
Interpretation
Uncertain
