Gene
KWMTBOMO04003 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012780
Annotation
putative_succinyl-CoA_synthetase_small_subunit_[Danaus_plexippus]
Full name
Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial
Alternative Name
Succinyl-CoA synthetase subunit alpha
Succinyl-coenzyme A synthetase alpha subunit 1
Succinyl-coenzyme A synthetase alpha subunit 1
Location in the cell
Mitochondrial Reliability : 2.656
Sequence
CDS
ATGGCTATGCCAGTCCGGGTTCTTTCTAAGTTCAAAGATGGACTAAAACTCGGCAATGTACGATTTGCATCCGGCAACCCTTATGCTGAAACCAGAAAAAATTTGATCCTTACAAGTGAAACAAAAGTGATTGTTCAGGGTTTCACTGGAAAGCAGGGTACCTTCCACAGCCAACAAGCCCTTGACTATGGTACTAAAGTTGTTGGAGGAGTGTCACCAAAGAAGGCTGGTACAGAACATCTTGGTAAGCCTGTGTTTGGTACAGTCAAAGAGGCAAAGGCAGGCACAGGAGCAACTGCCTCTGTTATTTATGTACCTCCCCCTGGAGCTGCAGCAGCCATTCTAGAAGCTATTGAGGCTGAAATGCCCCTCATTGTGTGCATTACAGAAGGTGTGCCACAACATGATATGGTGCGAGTAAAACATGCCCTCTTGAGACAAAACAAGTCTAGACTGGTTGGTCCTAACTGTCCTGGTATCATTGCTCCCGAGAAGTGTAAAATTGGTATCATGCCAGCTGCAGTGCACAAACGTGGTTGCATCGGAGTTGTATCGCGTTCTGGGACATTGACCTATGAAGCCTGTCATCAGACTACTATTACTGGCTTAGGCCAAACTTTGTGTGTCGGTATTGGAGGGGATCCCTTCAACGGAACAGACTTCATTGACTGCTTAGAAGTTTTCCTCAAGGATCCTGAGACCAAGGGCATCATCCTCATTGGTGAAATTGGTGGAAATGCTGAAGAATTAGCATCAGAATATTTGACTCAGTACAATACTGGTCAAAAAGCTAAGCCTGTGGTGTCGTTTATCGCCGGTTTGACGGCTCCCCCGGGTCGACGAATGGGTCACGCTGGTGCCATCATTTCGGGCGGCAAAGGAGGCGCCATGGACAAAATTAAGGCTTTAGAAAAAGCTAATGTAATCGTCACTCGGTCTCCGGCCAAAATGGGCGTGGAACTTCACAAAGAAATGAAACGATTAGAGATCATCTAA
Protein
MAMPVRVLSKFKDGLKLGNVRFASGNPYAETRKNLILTSETKVIVQGFTGKQGTFHSQQALDYGTKVVGGVSPKKAGTEHLGKPVFGTVKEAKAGTGATASVIYVPPPGAAAAILEAIEAEMPLIVCITEGVPQHDMVRVKHALLRQNKSRLVGPNCPGIIAPEKCKIGIMPAAVHKRGCIGVVSRSGTLTYEACHQTTITGLGQTLCVGIGGDPFNGTDFIDCLEVFLKDPETKGIILIGEIGGNAEELASEYLTQYNTGQKAKPVVSFIAGLTAPPGRRMGHAGAIISGGKGGAMDKIKALEKANVIVTRSPAKMGVELHKEMKRLEII
Summary
Description
Succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of either ATP or GTP and thus represents the only step of substrate-level phosphorylation in the TCA. The alpha subunit of the enzyme binds the substrates coenzyme A and phosphate, while succinate binding and specificity for either ATP or GTP is provided by different beta subunits.
Catalytic Activity
ATP + CoA + succinate = ADP + phosphate + succinyl-CoA
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
Subunit
Heterodimer of an alpha and a beta subunit. Different beta subunits determine nucleotide specificity. Together with an ATP-specific beta subunit, forms an ADP-forming succinyl-CoA synthetase (A-SCS). Together with a GTP-specific beta subunit forms a GDP-forming succinyl-CoA synthetase (G-SCS).
Similarity
Belongs to the succinate/malate CoA ligase alpha subunit family.
Keywords
Complete proteome
Ligase
Mitochondrion
Nucleotide-binding
Reference proteome
Transit peptide
Tricarboxylic acid cycle
Feature
chain Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial
Uniprot
H9JTB8
A0A3S2TGU8
A0A212EIE7
S4PML6
A0A194RKT2
A0A194Q5R0
+ More
E3UKP6 V5SHZ8 A0A2A4JTB3 A0A1L8E146 A0A0A1X3P7 Q1HR33 A0A023EPT6 A0A1Y9G8K1 A0A034W897 A0A0L0BQF0 A0A1I8Q4T6 A0A0K8VW67 A0A0K8TN56 A0A1I8JVE7 A0A1C7CZS7 A0A1I8JV33 A0A1I8JTE3 Q7PXA8 A0A1I8JUR8 A0A1L8EH99 A0A240PPQ4 A0A2C9GUM6 A0A1Y9HAB6 A0A1Y9IVV5 A0A2M4AC31 W5J816 A0A1Y9H2T8 W8BDF5 T1E7J0 A0A1I8M5I0 A0A1Q3G444 A0A2M3ZGR0 A0A240PKK7 V5GQY1 A0A1I8JW55 B3NFY4 A0A1J1IAG1 B4KY99 D3TRD7 A0A1B0AJC3 A0A1A9WLE1 A0A336K9C0 E2AUB5 B4N590 A0A0P4VP23 R4G324 A0A240PPA9 A0A336LUD5 B3M805 A0A069DSN1 A0A3B0KGQ0 B5DPQ3 B4GS06 A0A0M4ERI3 B4PH63 B4IZW2 A0A0A9XJW2 A0A0A9WL89 B4LDG6 W8C099 A0A182YBD5 A0A224XSF2 A0A0V0G6I8 A0A084VXV8 A0A0A9WN87 B4HTU6 A0A1W4W4Q4 B4QPY9 Q94522 A0A023F9Q6 A0A158P3V0 A0A0L7RGD4 E9J1L4 D6W7D6 A0A1B6MI96 A0A2J7QSL9 A0A1B6JRD9 A0A3L8DTP8 V9ID95 E0VG44 U5ELV2 A0A0L7LPY4 J3JZ14 A0A0P5JIU0 A0A0C9Q444 A0A151K0M7 A0A1B0GF61 A0A067QT32 A0A088ADV3 T1PCI4 A0A2A3E2W5 E2BNP4 R4WCU3 A0A1S3CX11
E3UKP6 V5SHZ8 A0A2A4JTB3 A0A1L8E146 A0A0A1X3P7 Q1HR33 A0A023EPT6 A0A1Y9G8K1 A0A034W897 A0A0L0BQF0 A0A1I8Q4T6 A0A0K8VW67 A0A0K8TN56 A0A1I8JVE7 A0A1C7CZS7 A0A1I8JV33 A0A1I8JTE3 Q7PXA8 A0A1I8JUR8 A0A1L8EH99 A0A240PPQ4 A0A2C9GUM6 A0A1Y9HAB6 A0A1Y9IVV5 A0A2M4AC31 W5J816 A0A1Y9H2T8 W8BDF5 T1E7J0 A0A1I8M5I0 A0A1Q3G444 A0A2M3ZGR0 A0A240PKK7 V5GQY1 A0A1I8JW55 B3NFY4 A0A1J1IAG1 B4KY99 D3TRD7 A0A1B0AJC3 A0A1A9WLE1 A0A336K9C0 E2AUB5 B4N590 A0A0P4VP23 R4G324 A0A240PPA9 A0A336LUD5 B3M805 A0A069DSN1 A0A3B0KGQ0 B5DPQ3 B4GS06 A0A0M4ERI3 B4PH63 B4IZW2 A0A0A9XJW2 A0A0A9WL89 B4LDG6 W8C099 A0A182YBD5 A0A224XSF2 A0A0V0G6I8 A0A084VXV8 A0A0A9WN87 B4HTU6 A0A1W4W4Q4 B4QPY9 Q94522 A0A023F9Q6 A0A158P3V0 A0A0L7RGD4 E9J1L4 D6W7D6 A0A1B6MI96 A0A2J7QSL9 A0A1B6JRD9 A0A3L8DTP8 V9ID95 E0VG44 U5ELV2 A0A0L7LPY4 J3JZ14 A0A0P5JIU0 A0A0C9Q444 A0A151K0M7 A0A1B0GF61 A0A067QT32 A0A088ADV3 T1PCI4 A0A2A3E2W5 E2BNP4 R4WCU3 A0A1S3CX11
EC Number
6.2.1.4
6.2.1.5
6.2.1.5
Pubmed
19121390
22118469
23622113
26354079
25830018
17204158
+ More
17510324 24945155 26483478 25348373 26108605 26369729 20966253 12364791 14747013 17210077 20920257 23761445 24495485 25315136 17994087 20353571 20798317 27129103 26334808 15632085 17550304 25401762 26823975 25244985 24438588 22936249 10731132 12537572 12537569 10071211 28017724 25474469 21347285 21282665 18362917 19820115 30249741 20566863 26227816 22516182 23537049 24845553 23691247
17510324 24945155 26483478 25348373 26108605 26369729 20966253 12364791 14747013 17210077 20920257 23761445 24495485 25315136 17994087 20353571 20798317 27129103 26334808 15632085 17550304 25401762 26823975 25244985 24438588 22936249 10731132 12537572 12537569 10071211 28017724 25474469 21347285 21282665 18362917 19820115 30249741 20566863 26227816 22516182 23537049 24845553 23691247
EMBL
BABH01004162
BABH01011634
BABH01011635
RSAL01000155
RVE45656.1
AGBW02014660
+ More
OWR41259.1 GAIX01003490 JAA89070.1 KQ460075 KPJ17940.1 KQ459582 KPI98735.1 HM449920 ADO33055.1 KF647633 AHB50495.1 NWSH01000620 PCG75251.1 GFDF01001792 JAV12292.1 GBXI01009014 JAD05278.1 DQ440261 CH477408 ABF18294.1 EAT41527.1 JXUM01044656 GAPW01002568 KQ561409 JAC11030.1 KXJ78673.1 GAKP01007171 GAKP01007170 JAC51781.1 JRES01001517 KNC22315.1 GDHF01009188 JAI43126.1 GDAI01002020 JAI15583.1 APCN01000834 AAAB01008987 EAA01194.2 GFDG01000774 JAV18025.1 AXCM01001181 AXCN02001518 GGFK01005022 MBW38343.1 ADMH02002105 ETN59000.1 GAMC01011533 GAMC01011532 JAB95023.1 GAMD01003002 JAA98588.1 GFDL01000472 JAV34573.1 GGFM01007006 MBW27757.1 GALX01001917 JAB66549.1 CH954178 EDV50813.1 CVRI01000047 CRK97203.1 CH933809 EDW17678.1 EZ423989 ADD20265.1 UFQS01000194 UFQT01000194 UFQT01000732 SSX01091.1 SSX21471.1 SSX26785.1 GL442815 EFN62972.1 CH964101 EDW79529.1 GDKW01002388 JAI54207.1 ACPB03016273 GAHY01002066 JAA75444.1 SSX01090.1 SSX21470.1 SSX26784.1 CH902618 EDV39913.1 GBGD01002148 JAC86741.1 OUUW01000009 SPP84916.1 CH379069 EDY73560.1 CH479188 EDW40541.1 CP012525 ALC45080.1 CM000159 EDW93300.1 CH916366 EDV96734.1 GBHO01024526 GBRD01000206 GDHC01021949 JAG19078.1 JAG65615.1 JAP96679.1 GBHO01034392 GBHO01034389 GBRD01017667 GBRD01014819 JAG09212.1 JAG09215.1 JAG48160.1 CH940647 EDW68904.1 GAMC01011531 GAMC01011530 JAB95024.1 GFTR01005056 JAW11370.1 GECL01002409 JAP03715.1 ATLV01018208 KE525224 KFB42802.1 GBHO01034390 JAG09214.1 CH480817 EDW50367.1 CM000363 CM002912 EDX09109.1 KMY97424.1 AE014296 AY089549 Y09067 GBBI01001054 JAC17658.1 ADTU01008562 KQ414596 KOC69915.1 GL767674 EFZ13224.1 KQ971307 EFA11129.1 GEBQ01004331 JAT35646.1 NEVH01011876 PNF31578.1 GECU01005917 JAT01790.1 QOIP01000005 RLU23138.1 JR038527 AEY58299.1 DS235131 EEB12350.1 GANO01001260 JAB58611.1 JTDY01000368 KOB77500.1 APGK01047228 APGK01047229 APGK01047230 BT128495 KB741077 AEE63452.1 ENN74242.1 GDIQ01197769 JAK53956.1 GBYB01008803 JAG78570.1 KQ981292 KYN43135.1 CCAG010012763 KK853313 KDR08649.1 KA646462 AFP61091.1 KZ288416 PBC26057.1 GL449438 EFN82692.1 AK417178 BAN20393.1
OWR41259.1 GAIX01003490 JAA89070.1 KQ460075 KPJ17940.1 KQ459582 KPI98735.1 HM449920 ADO33055.1 KF647633 AHB50495.1 NWSH01000620 PCG75251.1 GFDF01001792 JAV12292.1 GBXI01009014 JAD05278.1 DQ440261 CH477408 ABF18294.1 EAT41527.1 JXUM01044656 GAPW01002568 KQ561409 JAC11030.1 KXJ78673.1 GAKP01007171 GAKP01007170 JAC51781.1 JRES01001517 KNC22315.1 GDHF01009188 JAI43126.1 GDAI01002020 JAI15583.1 APCN01000834 AAAB01008987 EAA01194.2 GFDG01000774 JAV18025.1 AXCM01001181 AXCN02001518 GGFK01005022 MBW38343.1 ADMH02002105 ETN59000.1 GAMC01011533 GAMC01011532 JAB95023.1 GAMD01003002 JAA98588.1 GFDL01000472 JAV34573.1 GGFM01007006 MBW27757.1 GALX01001917 JAB66549.1 CH954178 EDV50813.1 CVRI01000047 CRK97203.1 CH933809 EDW17678.1 EZ423989 ADD20265.1 UFQS01000194 UFQT01000194 UFQT01000732 SSX01091.1 SSX21471.1 SSX26785.1 GL442815 EFN62972.1 CH964101 EDW79529.1 GDKW01002388 JAI54207.1 ACPB03016273 GAHY01002066 JAA75444.1 SSX01090.1 SSX21470.1 SSX26784.1 CH902618 EDV39913.1 GBGD01002148 JAC86741.1 OUUW01000009 SPP84916.1 CH379069 EDY73560.1 CH479188 EDW40541.1 CP012525 ALC45080.1 CM000159 EDW93300.1 CH916366 EDV96734.1 GBHO01024526 GBRD01000206 GDHC01021949 JAG19078.1 JAG65615.1 JAP96679.1 GBHO01034392 GBHO01034389 GBRD01017667 GBRD01014819 JAG09212.1 JAG09215.1 JAG48160.1 CH940647 EDW68904.1 GAMC01011531 GAMC01011530 JAB95024.1 GFTR01005056 JAW11370.1 GECL01002409 JAP03715.1 ATLV01018208 KE525224 KFB42802.1 GBHO01034390 JAG09214.1 CH480817 EDW50367.1 CM000363 CM002912 EDX09109.1 KMY97424.1 AE014296 AY089549 Y09067 GBBI01001054 JAC17658.1 ADTU01008562 KQ414596 KOC69915.1 GL767674 EFZ13224.1 KQ971307 EFA11129.1 GEBQ01004331 JAT35646.1 NEVH01011876 PNF31578.1 GECU01005917 JAT01790.1 QOIP01000005 RLU23138.1 JR038527 AEY58299.1 DS235131 EEB12350.1 GANO01001260 JAB58611.1 JTDY01000368 KOB77500.1 APGK01047228 APGK01047229 APGK01047230 BT128495 KB741077 AEE63452.1 ENN74242.1 GDIQ01197769 JAK53956.1 GBYB01008803 JAG78570.1 KQ981292 KYN43135.1 CCAG010012763 KK853313 KDR08649.1 KA646462 AFP61091.1 KZ288416 PBC26057.1 GL449438 EFN82692.1 AK417178 BAN20393.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000008820 UP000069940 UP000249989 UP000069272 UP000037069 UP000095300 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075900 UP000075885 UP000075883 UP000075886 UP000075920 UP000000673 UP000075884 UP000095301 UP000075880 UP000076407 UP000008711 UP000183832 UP000009192 UP000092445 UP000091820 UP000000311 UP000007798 UP000015103 UP000075881 UP000007801 UP000268350 UP000001819 UP000008744 UP000092553 UP000002282 UP000001070 UP000008792 UP000076408 UP000030765 UP000001292 UP000192221 UP000000304 UP000000803 UP000005205 UP000053825 UP000007266 UP000235965 UP000279307 UP000009046 UP000037510 UP000019118 UP000078541 UP000092444 UP000027135 UP000005203 UP000242457 UP000008237 UP000079169
UP000008820 UP000069940 UP000249989 UP000069272 UP000037069 UP000095300 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075900 UP000075885 UP000075883 UP000075886 UP000075920 UP000000673 UP000075884 UP000095301 UP000075880 UP000076407 UP000008711 UP000183832 UP000009192 UP000092445 UP000091820 UP000000311 UP000007798 UP000015103 UP000075881 UP000007801 UP000268350 UP000001819 UP000008744 UP000092553 UP000002282 UP000001070 UP000008792 UP000076408 UP000030765 UP000001292 UP000192221 UP000000304 UP000000803 UP000005205 UP000053825 UP000007266 UP000235965 UP000279307 UP000009046 UP000037510 UP000019118 UP000078541 UP000092444 UP000027135 UP000005203 UP000242457 UP000008237 UP000079169
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JTB8
A0A3S2TGU8
A0A212EIE7
S4PML6
A0A194RKT2
A0A194Q5R0
+ More
E3UKP6 V5SHZ8 A0A2A4JTB3 A0A1L8E146 A0A0A1X3P7 Q1HR33 A0A023EPT6 A0A1Y9G8K1 A0A034W897 A0A0L0BQF0 A0A1I8Q4T6 A0A0K8VW67 A0A0K8TN56 A0A1I8JVE7 A0A1C7CZS7 A0A1I8JV33 A0A1I8JTE3 Q7PXA8 A0A1I8JUR8 A0A1L8EH99 A0A240PPQ4 A0A2C9GUM6 A0A1Y9HAB6 A0A1Y9IVV5 A0A2M4AC31 W5J816 A0A1Y9H2T8 W8BDF5 T1E7J0 A0A1I8M5I0 A0A1Q3G444 A0A2M3ZGR0 A0A240PKK7 V5GQY1 A0A1I8JW55 B3NFY4 A0A1J1IAG1 B4KY99 D3TRD7 A0A1B0AJC3 A0A1A9WLE1 A0A336K9C0 E2AUB5 B4N590 A0A0P4VP23 R4G324 A0A240PPA9 A0A336LUD5 B3M805 A0A069DSN1 A0A3B0KGQ0 B5DPQ3 B4GS06 A0A0M4ERI3 B4PH63 B4IZW2 A0A0A9XJW2 A0A0A9WL89 B4LDG6 W8C099 A0A182YBD5 A0A224XSF2 A0A0V0G6I8 A0A084VXV8 A0A0A9WN87 B4HTU6 A0A1W4W4Q4 B4QPY9 Q94522 A0A023F9Q6 A0A158P3V0 A0A0L7RGD4 E9J1L4 D6W7D6 A0A1B6MI96 A0A2J7QSL9 A0A1B6JRD9 A0A3L8DTP8 V9ID95 E0VG44 U5ELV2 A0A0L7LPY4 J3JZ14 A0A0P5JIU0 A0A0C9Q444 A0A151K0M7 A0A1B0GF61 A0A067QT32 A0A088ADV3 T1PCI4 A0A2A3E2W5 E2BNP4 R4WCU3 A0A1S3CX11
E3UKP6 V5SHZ8 A0A2A4JTB3 A0A1L8E146 A0A0A1X3P7 Q1HR33 A0A023EPT6 A0A1Y9G8K1 A0A034W897 A0A0L0BQF0 A0A1I8Q4T6 A0A0K8VW67 A0A0K8TN56 A0A1I8JVE7 A0A1C7CZS7 A0A1I8JV33 A0A1I8JTE3 Q7PXA8 A0A1I8JUR8 A0A1L8EH99 A0A240PPQ4 A0A2C9GUM6 A0A1Y9HAB6 A0A1Y9IVV5 A0A2M4AC31 W5J816 A0A1Y9H2T8 W8BDF5 T1E7J0 A0A1I8M5I0 A0A1Q3G444 A0A2M3ZGR0 A0A240PKK7 V5GQY1 A0A1I8JW55 B3NFY4 A0A1J1IAG1 B4KY99 D3TRD7 A0A1B0AJC3 A0A1A9WLE1 A0A336K9C0 E2AUB5 B4N590 A0A0P4VP23 R4G324 A0A240PPA9 A0A336LUD5 B3M805 A0A069DSN1 A0A3B0KGQ0 B5DPQ3 B4GS06 A0A0M4ERI3 B4PH63 B4IZW2 A0A0A9XJW2 A0A0A9WL89 B4LDG6 W8C099 A0A182YBD5 A0A224XSF2 A0A0V0G6I8 A0A084VXV8 A0A0A9WN87 B4HTU6 A0A1W4W4Q4 B4QPY9 Q94522 A0A023F9Q6 A0A158P3V0 A0A0L7RGD4 E9J1L4 D6W7D6 A0A1B6MI96 A0A2J7QSL9 A0A1B6JRD9 A0A3L8DTP8 V9ID95 E0VG44 U5ELV2 A0A0L7LPY4 J3JZ14 A0A0P5JIU0 A0A0C9Q444 A0A151K0M7 A0A1B0GF61 A0A067QT32 A0A088ADV3 T1PCI4 A0A2A3E2W5 E2BNP4 R4WCU3 A0A1S3CX11
PDB
6G4Q
E-value=8.72438e-101,
Score=936
Ontologies
PATHWAY
GO
GO:0004775
GO:0004776
GO:0000166
GO:0005739
GO:0048037
GO:0006099
GO:0016874
GO:0006104
GO:0004550
GO:0006105
GO:0009142
GO:0005829
GO:0004778
GO:0016746
GO:0003735
GO:0019843
GO:0005840
GO:0006412
GO:0045244
GO:0003824
GO:0016021
GO:0016757
GO:0008152
GO:0008270
GO:0008892
GO:0006811
GO:0006032
GO:0004672
GO:0048384
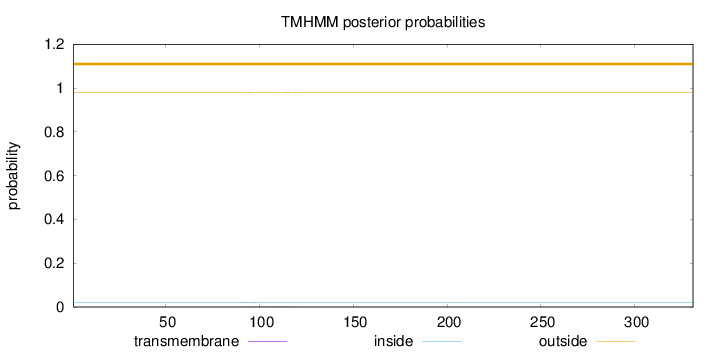
Topology
Subcellular location
Mitochondrion
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0368300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02115
outside
1 - 331
Population Genetic Test Statistics
Pi
231.087655
Theta
178.321308
Tajima's D
1.082502
CLR
0.079842
CSRT
0.68341582920854
Interpretation
Uncertain
