Pre Gene Modal
BGIBMGA010259
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_DDX43_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.13 Nuclear Reliability : 1.77
Sequence
CDS
ATGACTGATCGCGACGATGATGATTGGATTGAGAATGAAGAGGTAGTCGTACAGACACCGGCATCAAATGTACCTACACACAATACAAGGGTTTACCGACCAACGGCTAGTCGTGGAAATCGCGGCCGTAATACCGATTACAATTGGAGGTCTCGTGGTACAAACGAAGGCCAAAGTGGACGTTTTGACGATAGACGTTCTGGCTCTCGCCAAGAGGGTAGCAAAAAAGTTATTACCATACCTACCGATAAAGTAGGTAGGGTAATAGGTAAAGGTGGTTCAAAAATACGAGATCTGCAAGATGAATCTAAAGCAAAGATAAAGATAGGCGATTCCAGTGGTAATGAAACGGATGTAACCCTCTTCGGCACTGACGAAGCTATCGGTAAAGCTGAAACCATGGTGAATAACTTAATGGAAGACTACAAACCTGCTCCGAGGAAAGAATATGAAGAAGTGGAAACAAGTAACTCACAAGATTTTTTTAAGAAAAATGATGGTGGTGTCCAAGTAATTGACTGGGATAAACTCAATGCCTTTTATGATGAACAACAAAACAACAGATGGGATAAGCTGCCGCCAATTATCAAAGATTTTTACAATGAAGATCCATTGGTTAAGAATATGTCCAAAGCAGAAGTGGCTCATTGGCGTCAAGTCAATAATGACATACAAGTAAAAAGAACATTTGATGAAAAACCAGGCTTAAAACCCATACCAAATCCTGTTCAAACATTTGAACAAGCCTTTCAGCATTATCCAGAAATTTTAGATGAAATTTACAAGCAGGGATTCAAACAGCCATCACCAATACAATGCCAGGCATGGCCAATACTTCTTCGTGGTGATGATATGATAGGTATTGCACAGACAGGGACTGGTAAAACATTGGCTTTCTTATTACCTGCACTAATCCACATAGAAGGACAAACTATACCTCGAGATCAGAGGGAAGGCCCTACAGTACTCATCATGGCACCTACACGGGAGCTGGCACTTCAAATAGACAAAGAGGTAGCAAAGTATCAGTATAAAGGAATTAAATCAGTATGCTTATATGGTGGAGGTGACCGTAAAGAACAGATTAAAGTTGTTGCCAAGGGAGTTGATATAGTGATAGCTACTCCAGGCAGACTCAATGATTTGGTAATGGCCAGGCATTTGAATATCATAAATTTCTCATATATTGTTTTAGACGAGGCAGATAGGATGTTAGACATGGGTTTTGAGCCTCAAATCAGGAAATCACTATTTGATGTAAGACCTGATCGTCAAACAGTAATGACATCCGCAACATGGCCTACTGGAGTACGGCGCCTTGCTGAATCTTACATGAAAGATCCCATCCAAGTTAATGTTGGATCATTGGATTTAGCAGCAGTGCACACTGTCACACAACAAATTATGTTTGTAGAAGAAGATGACAAAGAAGCAGCTCTGTTTGACTTTTTGTATAATATGGAACCTACTGACAAGGTCATCATATTTTGTGGCAAGAAAGCAGTTGCTACCCACATTACTACTGAATTATGCCTTAAAGGTATACAATGCCAATCTTTACATGGCGATAGGGATCAAAGTGATCGTGAGGCAGCTTTGGAAGACATGCTAGATGGGACAGTCAACATTTTGATTGCCACTGATGTTGCATCTCGAGGAATTGATATAAAAGATTTGACACATGTCGTGAATTTTGATTTTCCAAGGCATATTGAAGAATACGTACACAGGGTGGGGAGAACAGGACGTGCAGGAAAAACAGGAATAGCTCTTTCTTTTGTTTCAAGAAACGATTGGGCTCATGCAGCTGAGCTCATAAAGATACTTGAAGAAGCAAATCAAGAAGTTCCTCAAGAGTTAGAGGAAATGGCGCAAAGATTTCAAGCTATGAAGATTAGAAGAGAGCAAGAGCCTGGTAGGGGGGGAGGACGTGGAGGTGGAAGAGGGGGTAGAGGGAGAGGCAGATGGTAA
Protein
MTDRDDDDWIENEEVVVQTPASNVPTHNTRVYRPTASRGNRGRNTDYNWRSRGTNEGQSGRFDDRRSGSRQEGSKKVITIPTDKVGRVIGKGGSKIRDLQDESKAKIKIGDSSGNETDVTLFGTDEAIGKAETMVNNLMEDYKPAPRKEYEEVETSNSQDFFKKNDGGVQVIDWDKLNAFYDEQQNNRWDKLPPIIKDFYNEDPLVKNMSKAEVAHWRQVNNDIQVKRTFDEKPGLKPIPNPVQTFEQAFQHYPEILDEIYKQGFKQPSPIQCQAWPILLRGDDMIGIAQTGTGKTLAFLLPALIHIEGQTIPRDQREGPTVLIMAPTRELALQIDKEVAKYQYKGIKSVCLYGGGDRKEQIKVVAKGVDIVIATPGRLNDLVMARHLNIINFSYIVLDEADRMLDMGFEPQIRKSLFDVRPDRQTVMTSATWPTGVRRLAESYMKDPIQVNVGSLDLAAVHTVTQQIMFVEEDDKEAALFDFLYNMEPTDKVIIFCGKKAVATHITTELCLKGIQCQSLHGDRDQSDREAALEDMLDGTVNILIATDVASRGIDIKDLTHVVNFDFPRHIEEYVHRVGRTGRAGKTGIALSFVSRNDWAHAAELIKILEEANQEVPQELEEMAQRFQAMKIRREQEPGRGGGRGGGRGGRGRGRW
Summary
Similarity
Belongs to the DEAD box helicase family.
Uniprot
H9JL58
A0A2W1BYR9
A0A2A4JV11
A0A212EIG6
A0A2H1X0W1
A0A0L7LQ25
+ More
A0A194RPJ3 A0A194Q168 E2A0G5 A0A0J7KDA9 A0A0L7QSF0 A0A151IFQ5 A0A1W4WFZ5 A0A067RR85 A0A195FKQ2 A0A151JBL1 A0A310SEX5 A0A154PPA8 A0A232EV41 F4WB06 K7J039 U4UHX0 N6UL26 A0A2A3E7G4 A0A151WN07 A0A1Y1M7X8 A0A2J7PMK7 A0A0M9ACK3 A0A158NRA5 E9IRZ5 A0A195BFE2 A0A087ZUZ1 A0A182FAR6 A0A139WEA2 E2BLC7 A0A2M4AP00 B0WEX6 A0A2M4AJZ0 A0A067ZU11 W5JGZ7 A0A182NCQ0 A0A182P3F4 A0A1Q3EYP6 T1JNX3 A0A182R8G9 U5ESK0 W8B453 A0A026W733 Q16T16 A0A2R5LID1 A0A1S4FQG3 A0A182JN43 A0A087U8V7 A0A336M3K0 A0A084WQ82 A0A1B6BXS7 A0A182MJI6 A0A182VAI7 A0A1S3K8C6 A0A023EVY9 A0A182TXX3 A0A1C7CZY3 A0A0P5US11 A7UT24 A0A182XF23 A0A0P6HUL9 A0A182YJP6 A0A0P5LFI3 A0A0P5TCY5 A0A0P5XVB0 A0A0P5FWQ4 A0A0P5NJ71 A0A0P6DX92 A0A0N8BTU9 A0A0N8E4X1 A0A0P5FYT7 A0A1B6IWX4 A0A182K4E1 A0A0P6CSN0 A0A0P5QJR2 A0A182W338 A0A1A9Z750 A0A0K8TLU5 A0A0N8BCK9 A0A0P5HKY3 A0A2M4BKM6 A0A0N8B4G5 A0A0P5NGN0 A0A0P5NRN7 A0A0P5NRP5 A0A182SDA7 A0A0P5K8L1 D3TME6 A0A1B0G2I2 A0A1B0ASM9 A0A1A9X5Z0 A0A1B0DN70 A0A1B6GVC3 A0A2L2Y758
A0A194RPJ3 A0A194Q168 E2A0G5 A0A0J7KDA9 A0A0L7QSF0 A0A151IFQ5 A0A1W4WFZ5 A0A067RR85 A0A195FKQ2 A0A151JBL1 A0A310SEX5 A0A154PPA8 A0A232EV41 F4WB06 K7J039 U4UHX0 N6UL26 A0A2A3E7G4 A0A151WN07 A0A1Y1M7X8 A0A2J7PMK7 A0A0M9ACK3 A0A158NRA5 E9IRZ5 A0A195BFE2 A0A087ZUZ1 A0A182FAR6 A0A139WEA2 E2BLC7 A0A2M4AP00 B0WEX6 A0A2M4AJZ0 A0A067ZU11 W5JGZ7 A0A182NCQ0 A0A182P3F4 A0A1Q3EYP6 T1JNX3 A0A182R8G9 U5ESK0 W8B453 A0A026W733 Q16T16 A0A2R5LID1 A0A1S4FQG3 A0A182JN43 A0A087U8V7 A0A336M3K0 A0A084WQ82 A0A1B6BXS7 A0A182MJI6 A0A182VAI7 A0A1S3K8C6 A0A023EVY9 A0A182TXX3 A0A1C7CZY3 A0A0P5US11 A7UT24 A0A182XF23 A0A0P6HUL9 A0A182YJP6 A0A0P5LFI3 A0A0P5TCY5 A0A0P5XVB0 A0A0P5FWQ4 A0A0P5NJ71 A0A0P6DX92 A0A0N8BTU9 A0A0N8E4X1 A0A0P5FYT7 A0A1B6IWX4 A0A182K4E1 A0A0P6CSN0 A0A0P5QJR2 A0A182W338 A0A1A9Z750 A0A0K8TLU5 A0A0N8BCK9 A0A0P5HKY3 A0A2M4BKM6 A0A0N8B4G5 A0A0P5NGN0 A0A0P5NRN7 A0A0P5NRP5 A0A182SDA7 A0A0P5K8L1 D3TME6 A0A1B0G2I2 A0A1B0ASM9 A0A1A9X5Z0 A0A1B0DN70 A0A1B6GVC3 A0A2L2Y758
Pubmed
EMBL
BABH01011634
KZ149940
PZC76923.1
NWSH01000620
PCG75250.1
AGBW02014660
+ More
OWR41260.1 ODYU01012550 SOQ58941.1 JTDY01000368 KOB77504.1 KQ460075 KPJ17941.1 KQ459582 KPI98734.1 GL435586 EFN73102.1 LBMM01009344 KMQ88194.1 KQ414766 KOC61391.1 KQ977771 KYM99994.1 KK852513 KDR22264.1 KQ981522 KYN40574.1 KQ979122 KYN22513.1 KQ762207 OAD56038.1 KQ435007 KZC13703.1 NNAY01002054 OXU22224.1 GL888056 EGI68626.1 KB632331 ERL92627.1 APGK01019271 APGK01019272 KB740098 ENN81391.1 KZ288345 PBC27640.1 KQ982922 KYQ49254.1 GEZM01042243 JAV80026.1 NEVH01023984 PNF17557.1 KQ435701 KOX80379.1 ADTU01023838 GL765283 EFZ16586.1 KQ976504 KYM82907.1 KQ971354 KYB26280.1 GL449017 EFN83479.1 GGFK01009188 MBW42509.1 DS231912 EDS25834.1 GGFK01007784 MBW41105.1 KF192311 AHY18681.1 ADMH02001552 ETN62089.1 GFDL01014621 JAV20424.1 JH431734 GANO01003214 JAB56657.1 GAMC01010625 JAB95930.1 KK107390 EZA51431.1 CH477661 EAT37615.1 GGLE01005072 MBY09198.1 KK118759 KFM73796.1 UFQT01000128 SSX20568.1 ATLV01025286 KE525379 KFB52376.1 GEDC01031478 GEDC01031476 JAS05820.1 JAS05822.1 AXCM01000114 GAPW01000306 JAC13292.1 GDIP01126381 LRGB01002227 JAL77333.1 KZS08342.1 AAAB01008898 EDO64138.2 GDIQ01014702 JAN80035.1 GDIQ01170359 JAK81366.1 GDIQ01093429 JAL58297.1 GDIP01066781 JAM36934.1 GDIQ01249335 JAK02390.1 GDIQ01207186 GDIQ01173496 GDIQ01171907 GDIQ01145468 GDIQ01097226 GDIQ01034709 JAL06258.1 GDIQ01072745 GDIQ01072744 JAN21992.1 GDIQ01208432 GDIQ01207187 GDIQ01145467 GDIQ01046455 JAL06259.1 GDIQ01061500 JAN33237.1 GDIQ01249336 JAK02389.1 GECU01016281 JAS91425.1 GDIQ01087890 JAN06847.1 GDIQ01134124 JAL17602.1 GDAI01002693 JAI14910.1 GDIQ01190987 JAK60738.1 GDIQ01228028 JAK23697.1 GGFJ01004464 MBW53605.1 GDIQ01213739 JAK37986.1 GDIQ01153105 JAK98620.1 GDIQ01138725 JAL13001.1 GDIQ01138726 JAL13000.1 GDIQ01213740 JAK37985.1 EZ422598 ADD18874.1 CCAG010018315 JXJN01002887 JXJN01002888 AJVK01017336 AJVK01017337 GECZ01003424 JAS66345.1 IAAA01013545 LAA03981.1
OWR41260.1 ODYU01012550 SOQ58941.1 JTDY01000368 KOB77504.1 KQ460075 KPJ17941.1 KQ459582 KPI98734.1 GL435586 EFN73102.1 LBMM01009344 KMQ88194.1 KQ414766 KOC61391.1 KQ977771 KYM99994.1 KK852513 KDR22264.1 KQ981522 KYN40574.1 KQ979122 KYN22513.1 KQ762207 OAD56038.1 KQ435007 KZC13703.1 NNAY01002054 OXU22224.1 GL888056 EGI68626.1 KB632331 ERL92627.1 APGK01019271 APGK01019272 KB740098 ENN81391.1 KZ288345 PBC27640.1 KQ982922 KYQ49254.1 GEZM01042243 JAV80026.1 NEVH01023984 PNF17557.1 KQ435701 KOX80379.1 ADTU01023838 GL765283 EFZ16586.1 KQ976504 KYM82907.1 KQ971354 KYB26280.1 GL449017 EFN83479.1 GGFK01009188 MBW42509.1 DS231912 EDS25834.1 GGFK01007784 MBW41105.1 KF192311 AHY18681.1 ADMH02001552 ETN62089.1 GFDL01014621 JAV20424.1 JH431734 GANO01003214 JAB56657.1 GAMC01010625 JAB95930.1 KK107390 EZA51431.1 CH477661 EAT37615.1 GGLE01005072 MBY09198.1 KK118759 KFM73796.1 UFQT01000128 SSX20568.1 ATLV01025286 KE525379 KFB52376.1 GEDC01031478 GEDC01031476 JAS05820.1 JAS05822.1 AXCM01000114 GAPW01000306 JAC13292.1 GDIP01126381 LRGB01002227 JAL77333.1 KZS08342.1 AAAB01008898 EDO64138.2 GDIQ01014702 JAN80035.1 GDIQ01170359 JAK81366.1 GDIQ01093429 JAL58297.1 GDIP01066781 JAM36934.1 GDIQ01249335 JAK02390.1 GDIQ01207186 GDIQ01173496 GDIQ01171907 GDIQ01145468 GDIQ01097226 GDIQ01034709 JAL06258.1 GDIQ01072745 GDIQ01072744 JAN21992.1 GDIQ01208432 GDIQ01207187 GDIQ01145467 GDIQ01046455 JAL06259.1 GDIQ01061500 JAN33237.1 GDIQ01249336 JAK02389.1 GECU01016281 JAS91425.1 GDIQ01087890 JAN06847.1 GDIQ01134124 JAL17602.1 GDAI01002693 JAI14910.1 GDIQ01190987 JAK60738.1 GDIQ01228028 JAK23697.1 GGFJ01004464 MBW53605.1 GDIQ01213739 JAK37986.1 GDIQ01153105 JAK98620.1 GDIQ01138725 JAL13001.1 GDIQ01138726 JAL13000.1 GDIQ01213740 JAK37985.1 EZ422598 ADD18874.1 CCAG010018315 JXJN01002887 JXJN01002888 AJVK01017336 AJVK01017337 GECZ01003424 JAS66345.1 IAAA01013545 LAA03981.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000000311 UP000036403 UP000053825 UP000078542 UP000192223 UP000027135 UP000078541 UP000078492 UP000076502 UP000215335 UP000007755 UP000002358 UP000030742 UP000019118 UP000242457 UP000075809 UP000235965 UP000053105 UP000005205 UP000078540 UP000005203 UP000069272 UP000007266 UP000008237 UP000002320 UP000000673 UP000075884 UP000075885 UP000075900 UP000053097 UP000008820 UP000075880 UP000054359 UP000030765 UP000075883 UP000075903 UP000085678 UP000075902 UP000075882 UP000076858 UP000007062 UP000076407 UP000076408 UP000075881 UP000075920 UP000092445 UP000075901 UP000092444 UP000092460 UP000092443 UP000092462
UP000000311 UP000036403 UP000053825 UP000078542 UP000192223 UP000027135 UP000078541 UP000078492 UP000076502 UP000215335 UP000007755 UP000002358 UP000030742 UP000019118 UP000242457 UP000075809 UP000235965 UP000053105 UP000005205 UP000078540 UP000005203 UP000069272 UP000007266 UP000008237 UP000002320 UP000000673 UP000075884 UP000075885 UP000075900 UP000053097 UP000008820 UP000075880 UP000054359 UP000030765 UP000075883 UP000075903 UP000085678 UP000075902 UP000075882 UP000076858 UP000007062 UP000076407 UP000076408 UP000075881 UP000075920 UP000092445 UP000075901 UP000092444 UP000092460 UP000092443 UP000092462
PRIDE
Pfam
Interpro
IPR001650
Helicase_C
+ More
IPR036612 KH_dom_type_1_sf
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR004088 KH_dom_type_1
IPR027417 P-loop_NTPase
IPR004087 KH_dom
IPR002591 Phosphodiest/P_Trfase
IPR017850 Alkaline_phosphatase_core_sf
IPR006116 2-5-oligoadenylate_synth_N
IPR036389 RNase_III_sf
IPR014720 dsRBD_dom
IPR000999 RNase_III_dom
IPR036085 PAZ_dom_sf
IPR003100 PAZ_dom
IPR036612 KH_dom_type_1_sf
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR004088 KH_dom_type_1
IPR027417 P-loop_NTPase
IPR004087 KH_dom
IPR002591 Phosphodiest/P_Trfase
IPR017850 Alkaline_phosphatase_core_sf
IPR006116 2-5-oligoadenylate_synth_N
IPR036389 RNase_III_sf
IPR014720 dsRBD_dom
IPR000999 RNase_III_dom
IPR036085 PAZ_dom_sf
IPR003100 PAZ_dom
SUPFAM
Gene 3D
ProteinModelPortal
H9JL58
A0A2W1BYR9
A0A2A4JV11
A0A212EIG6
A0A2H1X0W1
A0A0L7LQ25
+ More
A0A194RPJ3 A0A194Q168 E2A0G5 A0A0J7KDA9 A0A0L7QSF0 A0A151IFQ5 A0A1W4WFZ5 A0A067RR85 A0A195FKQ2 A0A151JBL1 A0A310SEX5 A0A154PPA8 A0A232EV41 F4WB06 K7J039 U4UHX0 N6UL26 A0A2A3E7G4 A0A151WN07 A0A1Y1M7X8 A0A2J7PMK7 A0A0M9ACK3 A0A158NRA5 E9IRZ5 A0A195BFE2 A0A087ZUZ1 A0A182FAR6 A0A139WEA2 E2BLC7 A0A2M4AP00 B0WEX6 A0A2M4AJZ0 A0A067ZU11 W5JGZ7 A0A182NCQ0 A0A182P3F4 A0A1Q3EYP6 T1JNX3 A0A182R8G9 U5ESK0 W8B453 A0A026W733 Q16T16 A0A2R5LID1 A0A1S4FQG3 A0A182JN43 A0A087U8V7 A0A336M3K0 A0A084WQ82 A0A1B6BXS7 A0A182MJI6 A0A182VAI7 A0A1S3K8C6 A0A023EVY9 A0A182TXX3 A0A1C7CZY3 A0A0P5US11 A7UT24 A0A182XF23 A0A0P6HUL9 A0A182YJP6 A0A0P5LFI3 A0A0P5TCY5 A0A0P5XVB0 A0A0P5FWQ4 A0A0P5NJ71 A0A0P6DX92 A0A0N8BTU9 A0A0N8E4X1 A0A0P5FYT7 A0A1B6IWX4 A0A182K4E1 A0A0P6CSN0 A0A0P5QJR2 A0A182W338 A0A1A9Z750 A0A0K8TLU5 A0A0N8BCK9 A0A0P5HKY3 A0A2M4BKM6 A0A0N8B4G5 A0A0P5NGN0 A0A0P5NRN7 A0A0P5NRP5 A0A182SDA7 A0A0P5K8L1 D3TME6 A0A1B0G2I2 A0A1B0ASM9 A0A1A9X5Z0 A0A1B0DN70 A0A1B6GVC3 A0A2L2Y758
A0A194RPJ3 A0A194Q168 E2A0G5 A0A0J7KDA9 A0A0L7QSF0 A0A151IFQ5 A0A1W4WFZ5 A0A067RR85 A0A195FKQ2 A0A151JBL1 A0A310SEX5 A0A154PPA8 A0A232EV41 F4WB06 K7J039 U4UHX0 N6UL26 A0A2A3E7G4 A0A151WN07 A0A1Y1M7X8 A0A2J7PMK7 A0A0M9ACK3 A0A158NRA5 E9IRZ5 A0A195BFE2 A0A087ZUZ1 A0A182FAR6 A0A139WEA2 E2BLC7 A0A2M4AP00 B0WEX6 A0A2M4AJZ0 A0A067ZU11 W5JGZ7 A0A182NCQ0 A0A182P3F4 A0A1Q3EYP6 T1JNX3 A0A182R8G9 U5ESK0 W8B453 A0A026W733 Q16T16 A0A2R5LID1 A0A1S4FQG3 A0A182JN43 A0A087U8V7 A0A336M3K0 A0A084WQ82 A0A1B6BXS7 A0A182MJI6 A0A182VAI7 A0A1S3K8C6 A0A023EVY9 A0A182TXX3 A0A1C7CZY3 A0A0P5US11 A7UT24 A0A182XF23 A0A0P6HUL9 A0A182YJP6 A0A0P5LFI3 A0A0P5TCY5 A0A0P5XVB0 A0A0P5FWQ4 A0A0P5NJ71 A0A0P6DX92 A0A0N8BTU9 A0A0N8E4X1 A0A0P5FYT7 A0A1B6IWX4 A0A182K4E1 A0A0P6CSN0 A0A0P5QJR2 A0A182W338 A0A1A9Z750 A0A0K8TLU5 A0A0N8BCK9 A0A0P5HKY3 A0A2M4BKM6 A0A0N8B4G5 A0A0P5NGN0 A0A0P5NRN7 A0A0P5NRP5 A0A182SDA7 A0A0P5K8L1 D3TME6 A0A1B0G2I2 A0A1B0ASM9 A0A1A9X5Z0 A0A1B0DN70 A0A1B6GVC3 A0A2L2Y758
PDB
3IUY
E-value=1.73543e-76,
Score=730
Ontologies
GO
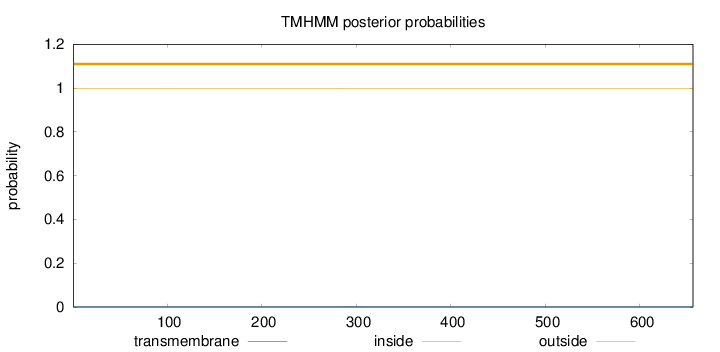
Topology
Length:
656
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01446
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00171
outside
1 - 656
Population Genetic Test Statistics
Pi
165.563714
Theta
144.057666
Tajima's D
-0.258912
CLR
311.851003
CSRT
0.295335233238338
Interpretation
Uncertain
