Gene
KWMTBOMO04001
Pre Gene Modal
BGIBMGA010121
Annotation
PREDICTED:_rho_GTPase-activating_protein_20_isoform_X3_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.459 Nuclear Reliability : 1.181
Sequence
CDS
ATGAGCAACTGGGCAAGGAGAATGCATCGCCGAGCAGCTACATTGAGTAATGTAGTCACGTCTTGTGGCCACCCGAATTCACTGCCCTATCCAAGAAGACTAGAAAAAGTCAAATTCGGCGTTCCACTTCTCGAAGTCTGCAAGGATGACATCCCGGGTCCCTTGCTCGTTCTGATATTGAAATTGAACAAGGAGGCACCGCTGCGCCGCGATGTGTTCCGCGCGCCGGGTCATCAGGGAGCCATGAAAAAATTAATTCATTTTCTTCAAACTGGCCGACTAGTCAATGTGGACAATTATTCTGTATATACCATTGCCAGTGTCTTGAAAAAGTTTCTCAGGAAGATACCAGGAGGCGTTTTCAGCCGCGATGGTGAAATGCAGCTCTTTGCAGCAGTTGAGCTGCCGGAGGAAAGGCAGGTCGATGAAATTCGCAGATTGCTAATGTCACTGCCCGTTTGCACGCAGCGATTATTGGTACTGTTGTTTGGTACGTTCCGGGTGATGGCTTCCAACGCTGACAAAGCGGGTACAGGCATGACATCGGAAGCTCTAGGCGTGTCGGTGGCGCCTTCGTTTTTCCACTCTTGCGTGTCGGACGGTAAAACGGCCACAATGGAGGATGTGCAGCGTTTCAAGGTGGCGACCCGTATAATGCGTCAGCTGATCGAGCAGTTCAGCGCCGGCGATCTGTTCGGGAGGGAGAATTACGAGTACTACGCTCGTGTGACGGGCCGCATTCTCCGCGTCAAGGACGAATGGATATGCTCGTTCAGGGATCCTCCGCCGCCGAGACATCGGAGAGACCAAGAGGCTTATGCTAGACAATTATCGCTTGAAGCAGAAAAGACATGGCTTCAATGCGAGTGCGACCGTTGGGGGAACAAGTTCAATCTCGAAGGTTTAACGAAATCCGAAGAGTGTCAGTCGATGCCCGCGCTGATGGGCGCTGTGGGCGGCGCCAACGCCGGCTCCGTGCTCGGCATCATACCGGAGCACAGCTTGCTCGACTCGTGCACCAGGCTGTCGATATCGCTCGAGGAAAGCATTTTTAAGATATCAGTCAGTTCGTCATCGCAATCGTCATGCAGTAACTCCAGTCCTCACATGACTCTGGAGGAGCTTCGGGCCATCAACAAGTACGCGGAGAGCACAAAGAGCCTCTCATATCTCCCTCAGTTGCAGGCGGCGAACGTCGCAGTACGCGGCGTCGGTCGCGGCGGTCGCGCGTGTGCTCGTGCCGCACGCTCGCGCACTCTGACACTGAGATCCCGCTCCCTCTCCAGCCTCGTAAAGTGA
Protein
MSNWARRMHRRAATLSNVVTSCGHPNSLPYPRRLEKVKFGVPLLEVCKDDIPGPLLVLILKLNKEAPLRRDVFRAPGHQGAMKKLIHFLQTGRLVNVDNYSVYTIASVLKKFLRKIPGGVFSRDGEMQLFAAVELPEERQVDEIRRLLMSLPVCTQRLLVLLFGTFRVMASNADKAGTGMTSEALGVSVAPSFFHSCVSDGKTATMEDVQRFKVATRIMRQLIEQFSAGDLFGRENYEYYARVTGRILRVKDEWICSFRDPPPPRHRRDQEAYARQLSLEAEKTWLQCECDRWGNKFNLEGLTKSEECQSMPALMGAVGGANAGSVLGIIPEHSLLDSCTRLSISLEESIFKISVSSSSQSSCSNSSPHMTLEELRAINKYAESTKSLSYLPQLQAANVAVRGVGRGGRACARAARSRTLTLRSRSLSSLVK
Summary
Cofactor
Fe cation
Similarity
Belongs to the fatty acid desaturase type 1 family.
Uniprot
A0A3S2NVJ0
A0A2W1BTF5
A0A194Q0A7
A0A212EIG3
H9JKS0
A0A1L8DNA1
+ More
A0A1S4G7V7 A0A084WMA9 A0A2P8XTY5 A0A0K8TNU7 A0A232ERZ1 A0A2J7Q547 K7IYW7 A0A2J7Q535 A0A067RG35 A0A2M4CZR4 A0A2M4ALL0 A0A0C9PQL5 E1ZUZ1 A0A1B6JEM9 A0A2M3ZCL1 A0A1B6GX54 A0A1B6JNS0 A0A1B6D582 A0A1B6LMA5 A0A151XHV5 A0A3L8DIX5 F4WHQ0 A0A151JUU3 A0A195B543 A0A158NM36 A0A151IBR7 E2C2U9 A0A151J0X8 U5EP11 T1HZ92 A0A0J7KD92 D2A378 A0A1B6BXZ7 A0A336MPF5 A0A0Q9XPX9 A0A1I8MGQ7 A0A0Q9WJP1 A0A0Q9WKC8 A0A0A9X8F7 A0A1I8Q2A5 A0A1B6IZ14 A0A1B6JSE3 B4N4W0 A0A0M4EFJ4 A0A1B0FLQ5 A0A0P9AKM9 A0A0R1E388 A0A0P8XUR9 B4PCI2 A0A0Q5UH14 A0A0R3P798 A0A146L3B2 A0A0A9X3B0 B3NI47 A0A0J9RW37 A0A0J9ULM8 M9MRW2 Q8IQM3 A0A1W4VAM2 B4HIH1 B4QL43 A0A1W4VNQ2 Q9VUR5 A0A2R7VTN4 N6UK12 A0A1I8MGQ3 A0A1Y1MS98 A0A0K8WIY9 A0A0A1XHG4 A0A034VSB3 A0A0A1WX73 A0A0L0C8F0 A0A0K8V8Q4 B4L0I6 B4LII8 A0A1I8Q2A1 A0A1B6CK66 A0A1I8Q292 A0A1I8MGQ5 B4J032 B3MAC7 A0A0R1E1P0 A0A0R1E3E0 A0A0P9C3Z3 A0A0J9RVP7 Q9VUR6 A0A0Q5U586 Q2M1D6 B7Z058 A0A0Q5UJD2 A0A1W4VN15 A0A1W4VMB5 U4UHW5 A0A026W615
A0A1S4G7V7 A0A084WMA9 A0A2P8XTY5 A0A0K8TNU7 A0A232ERZ1 A0A2J7Q547 K7IYW7 A0A2J7Q535 A0A067RG35 A0A2M4CZR4 A0A2M4ALL0 A0A0C9PQL5 E1ZUZ1 A0A1B6JEM9 A0A2M3ZCL1 A0A1B6GX54 A0A1B6JNS0 A0A1B6D582 A0A1B6LMA5 A0A151XHV5 A0A3L8DIX5 F4WHQ0 A0A151JUU3 A0A195B543 A0A158NM36 A0A151IBR7 E2C2U9 A0A151J0X8 U5EP11 T1HZ92 A0A0J7KD92 D2A378 A0A1B6BXZ7 A0A336MPF5 A0A0Q9XPX9 A0A1I8MGQ7 A0A0Q9WJP1 A0A0Q9WKC8 A0A0A9X8F7 A0A1I8Q2A5 A0A1B6IZ14 A0A1B6JSE3 B4N4W0 A0A0M4EFJ4 A0A1B0FLQ5 A0A0P9AKM9 A0A0R1E388 A0A0P8XUR9 B4PCI2 A0A0Q5UH14 A0A0R3P798 A0A146L3B2 A0A0A9X3B0 B3NI47 A0A0J9RW37 A0A0J9ULM8 M9MRW2 Q8IQM3 A0A1W4VAM2 B4HIH1 B4QL43 A0A1W4VNQ2 Q9VUR5 A0A2R7VTN4 N6UK12 A0A1I8MGQ3 A0A1Y1MS98 A0A0K8WIY9 A0A0A1XHG4 A0A034VSB3 A0A0A1WX73 A0A0L0C8F0 A0A0K8V8Q4 B4L0I6 B4LII8 A0A1I8Q2A1 A0A1B6CK66 A0A1I8Q292 A0A1I8MGQ5 B4J032 B3MAC7 A0A0R1E1P0 A0A0R1E3E0 A0A0P9C3Z3 A0A0J9RVP7 Q9VUR6 A0A0Q5U586 Q2M1D6 B7Z058 A0A0Q5UJD2 A0A1W4VN15 A0A1W4VMB5 U4UHW5 A0A026W615
Pubmed
28756777
26354079
22118469
19121390
24438588
29403074
+ More
26369729 28648823 20075255 24845553 20798317 30249741 21719571 21347285 18362917 19820115 17994087 25315136 25401762 26823975 17550304 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 28004739 25830018 25348373 26108605 18057021 23185243 24508170
26369729 28648823 20075255 24845553 20798317 30249741 21719571 21347285 18362917 19820115 17994087 25315136 25401762 26823975 17550304 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 28004739 25830018 25348373 26108605 18057021 23185243 24508170
EMBL
RSAL01000155
RVE45655.1
KZ149940
PZC76924.1
KQ459582
KPI98733.1
+ More
AGBW02014660 OWR41261.1 BABH01011632 BABH01011633 BABH01011634 GFDF01006239 JAV07845.1 ATLV01024395 KE525351 KFB51353.1 PYGN01001356 PSN35460.1 GDAI01002023 JAI15580.1 NNAY01002541 OXU21102.1 NEVH01018372 PNF23700.1 PNF23699.1 KK852497 KDR22717.1 GGFL01006543 MBW70721.1 GGFK01008301 MBW41622.1 GBYB01003518 JAG73285.1 GL434335 EFN74989.1 GECU01009955 JAS97751.1 GGFM01005491 MBW26242.1 GECZ01002763 JAS67006.1 GECU01006877 JAT00830.1 GEDC01016437 JAS20861.1 GEBQ01015353 JAT24624.1 KQ982109 KYQ59986.1 QOIP01000007 RLU20384.1 GL888167 EGI66257.1 KQ981730 KYN36608.1 KQ976604 KYM79387.1 ADTU01020219 KQ978079 KYM97167.1 GL452203 EFN77784.1 KQ980586 KYN15245.1 GANO01004871 JAB55000.1 ACPB03024846 LBMM01009408 KMQ88159.1 KQ971338 EFA01951.2 GEDC01031463 JAS05835.1 UFQS01001969 UFQT01001969 SSX12794.1 SSX32236.1 CH933809 KRG06519.1 CH940647 KRF85100.1 KRF85098.1 GBHO01030230 GBHO01030224 GBHO01029128 GBHO01029126 GBHO01029124 GBHO01027815 GBHO01027806 GBHO01027805 GBHO01023999 GDHC01001734 JAG13374.1 JAG13380.1 JAG14476.1 JAG14478.1 JAG14480.1 JAG15789.1 JAG15798.1 JAG15799.1 JAG19605.1 JAQ16895.1 GECU01015543 JAS92163.1 GECU01005607 JAT02100.1 CH964101 EDW79399.1 CP012525 ALC44734.1 CCAG010023511 CH902618 KPU78421.1 CM000159 KRK02098.1 KPU78423.1 EDW94892.2 CH954178 KQS44115.1 CH379069 KRT07493.1 GDHC01015676 JAQ02953.1 GBHO01038426 GBHO01038422 GBHO01038415 GBHO01030225 GBHO01029125 GBHO01029121 GBHO01029120 GBHO01027813 GBHO01024000 JAG05178.1 JAG05182.1 JAG05189.1 JAG13379.1 JAG14479.1 JAG14483.1 JAG14484.1 JAG15791.1 JAG19604.1 EDV52133.1 CM002912 KMY99787.1 KMY99785.1 AE014296 ADV37548.1 AAN11778.1 CH480815 EDW41601.1 CM000363 EDX10568.1 AY061070 AAF49610.2 AAL28618.1 KK854082 PTY10883.1 APGK01030244 APGK01030245 APGK01030246 KB740764 ENN79022.1 GEZM01024130 GEZM01024129 JAV87948.1 GDHF01001257 JAI51057.1 GBXI01004339 JAD09953.1 GAKP01013598 JAC45354.1 GBXI01011274 JAD03018.1 JRES01000760 KNC28520.1 GDHF01017088 JAI35226.1 EDW19155.1 KRG06517.1 KRG06518.1 EDW70775.1 GEDC01023451 JAS13847.1 CH916366 EDV97825.1 EDV40178.2 KRK02099.1 KRK02100.1 KPU78424.1 KPU78425.1 KMY99788.1 AAF49609.2 KQS44117.1 EAL30638.2 KRT07491.1 KRT07494.1 ACL83311.1 KQS44118.1 KB632308 ERL91933.1 KK107390 EZA51448.1
AGBW02014660 OWR41261.1 BABH01011632 BABH01011633 BABH01011634 GFDF01006239 JAV07845.1 ATLV01024395 KE525351 KFB51353.1 PYGN01001356 PSN35460.1 GDAI01002023 JAI15580.1 NNAY01002541 OXU21102.1 NEVH01018372 PNF23700.1 PNF23699.1 KK852497 KDR22717.1 GGFL01006543 MBW70721.1 GGFK01008301 MBW41622.1 GBYB01003518 JAG73285.1 GL434335 EFN74989.1 GECU01009955 JAS97751.1 GGFM01005491 MBW26242.1 GECZ01002763 JAS67006.1 GECU01006877 JAT00830.1 GEDC01016437 JAS20861.1 GEBQ01015353 JAT24624.1 KQ982109 KYQ59986.1 QOIP01000007 RLU20384.1 GL888167 EGI66257.1 KQ981730 KYN36608.1 KQ976604 KYM79387.1 ADTU01020219 KQ978079 KYM97167.1 GL452203 EFN77784.1 KQ980586 KYN15245.1 GANO01004871 JAB55000.1 ACPB03024846 LBMM01009408 KMQ88159.1 KQ971338 EFA01951.2 GEDC01031463 JAS05835.1 UFQS01001969 UFQT01001969 SSX12794.1 SSX32236.1 CH933809 KRG06519.1 CH940647 KRF85100.1 KRF85098.1 GBHO01030230 GBHO01030224 GBHO01029128 GBHO01029126 GBHO01029124 GBHO01027815 GBHO01027806 GBHO01027805 GBHO01023999 GDHC01001734 JAG13374.1 JAG13380.1 JAG14476.1 JAG14478.1 JAG14480.1 JAG15789.1 JAG15798.1 JAG15799.1 JAG19605.1 JAQ16895.1 GECU01015543 JAS92163.1 GECU01005607 JAT02100.1 CH964101 EDW79399.1 CP012525 ALC44734.1 CCAG010023511 CH902618 KPU78421.1 CM000159 KRK02098.1 KPU78423.1 EDW94892.2 CH954178 KQS44115.1 CH379069 KRT07493.1 GDHC01015676 JAQ02953.1 GBHO01038426 GBHO01038422 GBHO01038415 GBHO01030225 GBHO01029125 GBHO01029121 GBHO01029120 GBHO01027813 GBHO01024000 JAG05178.1 JAG05182.1 JAG05189.1 JAG13379.1 JAG14479.1 JAG14483.1 JAG14484.1 JAG15791.1 JAG19604.1 EDV52133.1 CM002912 KMY99787.1 KMY99785.1 AE014296 ADV37548.1 AAN11778.1 CH480815 EDW41601.1 CM000363 EDX10568.1 AY061070 AAF49610.2 AAL28618.1 KK854082 PTY10883.1 APGK01030244 APGK01030245 APGK01030246 KB740764 ENN79022.1 GEZM01024130 GEZM01024129 JAV87948.1 GDHF01001257 JAI51057.1 GBXI01004339 JAD09953.1 GAKP01013598 JAC45354.1 GBXI01011274 JAD03018.1 JRES01000760 KNC28520.1 GDHF01017088 JAI35226.1 EDW19155.1 KRG06517.1 KRG06518.1 EDW70775.1 GEDC01023451 JAS13847.1 CH916366 EDV97825.1 EDV40178.2 KRK02099.1 KRK02100.1 KPU78424.1 KPU78425.1 KMY99788.1 AAF49609.2 KQS44117.1 EAL30638.2 KRT07491.1 KRT07494.1 ACL83311.1 KQS44118.1 KB632308 ERL91933.1 KK107390 EZA51448.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000005204
UP000030765
UP000245037
+ More
UP000215335 UP000235965 UP000002358 UP000027135 UP000000311 UP000075809 UP000279307 UP000007755 UP000078541 UP000078540 UP000005205 UP000078542 UP000008237 UP000078492 UP000015103 UP000036403 UP000007266 UP000009192 UP000095301 UP000008792 UP000095300 UP000007798 UP000092553 UP000092444 UP000007801 UP000002282 UP000008711 UP000001819 UP000000803 UP000192221 UP000001292 UP000000304 UP000019118 UP000037069 UP000001070 UP000030742 UP000053097
UP000215335 UP000235965 UP000002358 UP000027135 UP000000311 UP000075809 UP000279307 UP000007755 UP000078541 UP000078540 UP000005205 UP000078542 UP000008237 UP000078492 UP000015103 UP000036403 UP000007266 UP000009192 UP000095301 UP000008792 UP000095300 UP000007798 UP000092553 UP000092444 UP000007801 UP000002282 UP000008711 UP000001819 UP000000803 UP000192221 UP000001292 UP000000304 UP000019118 UP000037069 UP000001070 UP000030742 UP000053097
Interpro
SUPFAM
SSF48350
SSF48350
Gene 3D
ProteinModelPortal
A0A3S2NVJ0
A0A2W1BTF5
A0A194Q0A7
A0A212EIG3
H9JKS0
A0A1L8DNA1
+ More
A0A1S4G7V7 A0A084WMA9 A0A2P8XTY5 A0A0K8TNU7 A0A232ERZ1 A0A2J7Q547 K7IYW7 A0A2J7Q535 A0A067RG35 A0A2M4CZR4 A0A2M4ALL0 A0A0C9PQL5 E1ZUZ1 A0A1B6JEM9 A0A2M3ZCL1 A0A1B6GX54 A0A1B6JNS0 A0A1B6D582 A0A1B6LMA5 A0A151XHV5 A0A3L8DIX5 F4WHQ0 A0A151JUU3 A0A195B543 A0A158NM36 A0A151IBR7 E2C2U9 A0A151J0X8 U5EP11 T1HZ92 A0A0J7KD92 D2A378 A0A1B6BXZ7 A0A336MPF5 A0A0Q9XPX9 A0A1I8MGQ7 A0A0Q9WJP1 A0A0Q9WKC8 A0A0A9X8F7 A0A1I8Q2A5 A0A1B6IZ14 A0A1B6JSE3 B4N4W0 A0A0M4EFJ4 A0A1B0FLQ5 A0A0P9AKM9 A0A0R1E388 A0A0P8XUR9 B4PCI2 A0A0Q5UH14 A0A0R3P798 A0A146L3B2 A0A0A9X3B0 B3NI47 A0A0J9RW37 A0A0J9ULM8 M9MRW2 Q8IQM3 A0A1W4VAM2 B4HIH1 B4QL43 A0A1W4VNQ2 Q9VUR5 A0A2R7VTN4 N6UK12 A0A1I8MGQ3 A0A1Y1MS98 A0A0K8WIY9 A0A0A1XHG4 A0A034VSB3 A0A0A1WX73 A0A0L0C8F0 A0A0K8V8Q4 B4L0I6 B4LII8 A0A1I8Q2A1 A0A1B6CK66 A0A1I8Q292 A0A1I8MGQ5 B4J032 B3MAC7 A0A0R1E1P0 A0A0R1E3E0 A0A0P9C3Z3 A0A0J9RVP7 Q9VUR6 A0A0Q5U586 Q2M1D6 B7Z058 A0A0Q5UJD2 A0A1W4VN15 A0A1W4VMB5 U4UHW5 A0A026W615
A0A1S4G7V7 A0A084WMA9 A0A2P8XTY5 A0A0K8TNU7 A0A232ERZ1 A0A2J7Q547 K7IYW7 A0A2J7Q535 A0A067RG35 A0A2M4CZR4 A0A2M4ALL0 A0A0C9PQL5 E1ZUZ1 A0A1B6JEM9 A0A2M3ZCL1 A0A1B6GX54 A0A1B6JNS0 A0A1B6D582 A0A1B6LMA5 A0A151XHV5 A0A3L8DIX5 F4WHQ0 A0A151JUU3 A0A195B543 A0A158NM36 A0A151IBR7 E2C2U9 A0A151J0X8 U5EP11 T1HZ92 A0A0J7KD92 D2A378 A0A1B6BXZ7 A0A336MPF5 A0A0Q9XPX9 A0A1I8MGQ7 A0A0Q9WJP1 A0A0Q9WKC8 A0A0A9X8F7 A0A1I8Q2A5 A0A1B6IZ14 A0A1B6JSE3 B4N4W0 A0A0M4EFJ4 A0A1B0FLQ5 A0A0P9AKM9 A0A0R1E388 A0A0P8XUR9 B4PCI2 A0A0Q5UH14 A0A0R3P798 A0A146L3B2 A0A0A9X3B0 B3NI47 A0A0J9RW37 A0A0J9ULM8 M9MRW2 Q8IQM3 A0A1W4VAM2 B4HIH1 B4QL43 A0A1W4VNQ2 Q9VUR5 A0A2R7VTN4 N6UK12 A0A1I8MGQ3 A0A1Y1MS98 A0A0K8WIY9 A0A0A1XHG4 A0A034VSB3 A0A0A1WX73 A0A0L0C8F0 A0A0K8V8Q4 B4L0I6 B4LII8 A0A1I8Q2A1 A0A1B6CK66 A0A1I8Q292 A0A1I8MGQ5 B4J032 B3MAC7 A0A0R1E1P0 A0A0R1E3E0 A0A0P9C3Z3 A0A0J9RVP7 Q9VUR6 A0A0Q5U586 Q2M1D6 B7Z058 A0A0Q5UJD2 A0A1W4VN15 A0A1W4VMB5 U4UHW5 A0A026W615
PDB
3MSX
E-value=2.30324e-18,
Score=227
Ontologies
GO
Topology
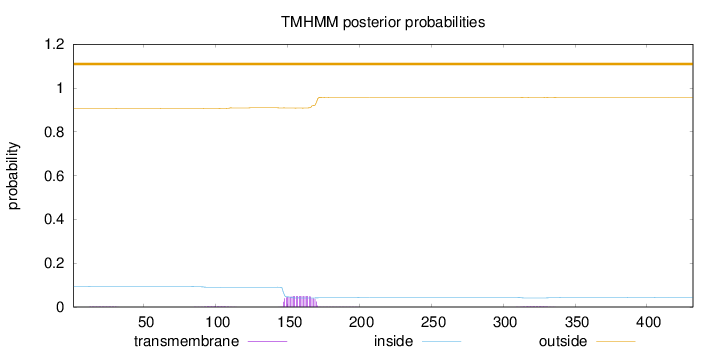
Length:
432
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.2495
Exp number, first 60 AAs:
0.0328
Total prob of N-in:
0.09482
outside
1 - 432
Population Genetic Test Statistics
Pi
248.569256
Theta
178.576656
Tajima's D
0.899126
CLR
0.450834
CSRT
0.635168241587921
Interpretation
Uncertain
