Gene
KWMTBOMO03994
Pre Gene Modal
BGIBMGA010124
Annotation
PREDICTED:_semaphorin-5B_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.428
Sequence
CDS
ATGGATAACGAATTGTACGCGATGCCGGCGTACAGCCAGTTCGATGAGGTGGCTTCGTTTGTATCAATGCCAAACGAGACCCTCAGCGTCGGGAGCATCGTCGGCTGTGTTATCGCTGCCTTTGTTATGGGCTGTCTACTCTGCTTGTCCCTTGTCATATTCTGCCAGCGCCGCGGCACGAGGATGCCATGGAAGAAACAGCATAGAGTTCCGTCAAGTCCTCACTACATCACAGCTAAACAAAACAGCTATGTGACCGTGCCTTTAAAGGAAGTGCCTCGTAAAGCAAAACGCCAGCCATCTTTTACTGGTATCGGAGGAGGCAGTGGGATTGTACTATCAAAGAGCAACAACATATCGAATGCTAATAACCACAATAATTCAGTTGCGACGCCCAAGCTTTACCCTAAGGCTATAGCAAATGAATACGATTCCATGGGAACGATAAGGAGGCACTCAAATCAACCGAACAACAAAAATAATTTGGATGTCGAAGAGGATAGATTTTATTAA
Protein
MDNELYAMPAYSQFDEVASFVSMPNETLSVGSIVGCVIAAFVMGCLLCLSLVIFCQRRGTRMPWKKQHRVPSSPHYITAKQNSYVTVPLKEVPRKAKRQPSFTGIGGGSGIVLSKSNNISNANNHNNSVATPKLYPKAIANEYDSMGTIRRHSNQPNNKNNLDVEEDRFY
Summary
Uniprot
H9JKS3
A0A2W1BPF3
A0A2A4JXU7
A0A2H1VVR5
A0A3S2NQ82
A0A194PZH7
+ More
A0A194RJE1 A0A0L7LPM2 A0A2A3ECC7 A0A3L8DQJ7 A0A026WC50 A0A0L7QKV9 A0A088AFH5 A0A154PGZ2 A0A310S489 E2B8I4 E9J776 A0A0N0BJV7 F4W964 A0A195EEJ4 A0A158P0G6 A0A195B4Z3 A0A151IFA6 A0A195F3E0 A0A0A9XBJ1 A0A1B6K4N4 A0A2P8Y884 A0A1B6DGV9 A0A1B6CGE7 A0A1B6LK84 A0A1B6ICN0 A0A0C9RD74 A0A1B6FUX1 A0A2J7RLU3 A0A067RM86 A0A0C9QVG8 A0A2H8TE71 K7J4N7 A0A3Q0ISN4 J9K1H2 A0A0T6BC38
A0A194RJE1 A0A0L7LPM2 A0A2A3ECC7 A0A3L8DQJ7 A0A026WC50 A0A0L7QKV9 A0A088AFH5 A0A154PGZ2 A0A310S489 E2B8I4 E9J776 A0A0N0BJV7 F4W964 A0A195EEJ4 A0A158P0G6 A0A195B4Z3 A0A151IFA6 A0A195F3E0 A0A0A9XBJ1 A0A1B6K4N4 A0A2P8Y884 A0A1B6DGV9 A0A1B6CGE7 A0A1B6LK84 A0A1B6ICN0 A0A0C9RD74 A0A1B6FUX1 A0A2J7RLU3 A0A067RM86 A0A0C9QVG8 A0A2H8TE71 K7J4N7 A0A3Q0ISN4 J9K1H2 A0A0T6BC38
Pubmed
EMBL
BABH01011623
KZ149940
PZC76932.1
NWSH01000377
PCG76845.1
ODYU01004694
+ More
SOQ44846.1 RSAL01000155 RVE45650.1 KQ459582 KPI98726.1 KQ460075 KPJ17948.1 JTDY01000402 KOB77349.1 KZ288287 PBC29378.1 QOIP01000005 RLU22473.1 KK107293 EZA53246.1 KQ414940 KOC59161.1 KQ434902 KZC11082.1 KQ780658 OAD51942.1 GL446332 EFN88018.1 GL768421 EFZ11332.1 KQ435710 KOX79712.1 GL888002 EGI69246.1 KQ979039 KYN23269.1 ADTU01000987 ADTU01000988 ADTU01000989 ADTU01000990 KQ976598 KYM79566.1 KQ977800 KYM99564.1 KQ981855 KYN34901.1 GBHO01025529 GBHO01025527 GBRD01010183 GDHC01009373 JAG18075.1 JAG18077.1 JAG55641.1 JAQ09256.1 GECU01001563 JAT06144.1 PYGN01000812 PSN40455.1 GEDC01012369 GEDC01000635 JAS24929.1 JAS36663.1 GEDC01024893 JAS12405.1 GEBQ01015953 JAT24024.1 GECU01035195 GECU01030805 GECU01023044 JAS72511.1 JAS76901.1 JAS84662.1 GBYB01004881 JAG74648.1 GECZ01015759 GECZ01007941 JAS54010.1 JAS61828.1 NEVH01002683 PNF41802.1 KK852543 KDR21695.1 GBYB01007714 GBYB01013784 JAG77481.1 JAG83551.1 GFXV01000591 MBW12396.1 AAZX01003821 AAZX01010313 AAZX01010399 AAZX01010407 AAZX01012808 ABLF02027390 LJIG01002177 KRT84763.1
SOQ44846.1 RSAL01000155 RVE45650.1 KQ459582 KPI98726.1 KQ460075 KPJ17948.1 JTDY01000402 KOB77349.1 KZ288287 PBC29378.1 QOIP01000005 RLU22473.1 KK107293 EZA53246.1 KQ414940 KOC59161.1 KQ434902 KZC11082.1 KQ780658 OAD51942.1 GL446332 EFN88018.1 GL768421 EFZ11332.1 KQ435710 KOX79712.1 GL888002 EGI69246.1 KQ979039 KYN23269.1 ADTU01000987 ADTU01000988 ADTU01000989 ADTU01000990 KQ976598 KYM79566.1 KQ977800 KYM99564.1 KQ981855 KYN34901.1 GBHO01025529 GBHO01025527 GBRD01010183 GDHC01009373 JAG18075.1 JAG18077.1 JAG55641.1 JAQ09256.1 GECU01001563 JAT06144.1 PYGN01000812 PSN40455.1 GEDC01012369 GEDC01000635 JAS24929.1 JAS36663.1 GEDC01024893 JAS12405.1 GEBQ01015953 JAT24024.1 GECU01035195 GECU01030805 GECU01023044 JAS72511.1 JAS76901.1 JAS84662.1 GBYB01004881 JAG74648.1 GECZ01015759 GECZ01007941 JAS54010.1 JAS61828.1 NEVH01002683 PNF41802.1 KK852543 KDR21695.1 GBYB01007714 GBYB01013784 JAG77481.1 JAG83551.1 GFXV01000591 MBW12396.1 AAZX01003821 AAZX01010313 AAZX01010399 AAZX01010407 AAZX01012808 ABLF02027390 LJIG01002177 KRT84763.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000037510
+ More
UP000242457 UP000279307 UP000053097 UP000053825 UP000005203 UP000076502 UP000008237 UP000053105 UP000007755 UP000078492 UP000005205 UP000078540 UP000078542 UP000078541 UP000245037 UP000235965 UP000027135 UP000002358 UP000079169 UP000007819
UP000242457 UP000279307 UP000053097 UP000053825 UP000005203 UP000076502 UP000008237 UP000053105 UP000007755 UP000078492 UP000005205 UP000078540 UP000078542 UP000078541 UP000245037 UP000235965 UP000027135 UP000002358 UP000079169 UP000007819
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JKS3
A0A2W1BPF3
A0A2A4JXU7
A0A2H1VVR5
A0A3S2NQ82
A0A194PZH7
+ More
A0A194RJE1 A0A0L7LPM2 A0A2A3ECC7 A0A3L8DQJ7 A0A026WC50 A0A0L7QKV9 A0A088AFH5 A0A154PGZ2 A0A310S489 E2B8I4 E9J776 A0A0N0BJV7 F4W964 A0A195EEJ4 A0A158P0G6 A0A195B4Z3 A0A151IFA6 A0A195F3E0 A0A0A9XBJ1 A0A1B6K4N4 A0A2P8Y884 A0A1B6DGV9 A0A1B6CGE7 A0A1B6LK84 A0A1B6ICN0 A0A0C9RD74 A0A1B6FUX1 A0A2J7RLU3 A0A067RM86 A0A0C9QVG8 A0A2H8TE71 K7J4N7 A0A3Q0ISN4 J9K1H2 A0A0T6BC38
A0A194RJE1 A0A0L7LPM2 A0A2A3ECC7 A0A3L8DQJ7 A0A026WC50 A0A0L7QKV9 A0A088AFH5 A0A154PGZ2 A0A310S489 E2B8I4 E9J776 A0A0N0BJV7 F4W964 A0A195EEJ4 A0A158P0G6 A0A195B4Z3 A0A151IFA6 A0A195F3E0 A0A0A9XBJ1 A0A1B6K4N4 A0A2P8Y884 A0A1B6DGV9 A0A1B6CGE7 A0A1B6LK84 A0A1B6ICN0 A0A0C9RD74 A0A1B6FUX1 A0A2J7RLU3 A0A067RM86 A0A0C9QVG8 A0A2H8TE71 K7J4N7 A0A3Q0ISN4 J9K1H2 A0A0T6BC38
Ontologies
GO
PANTHER
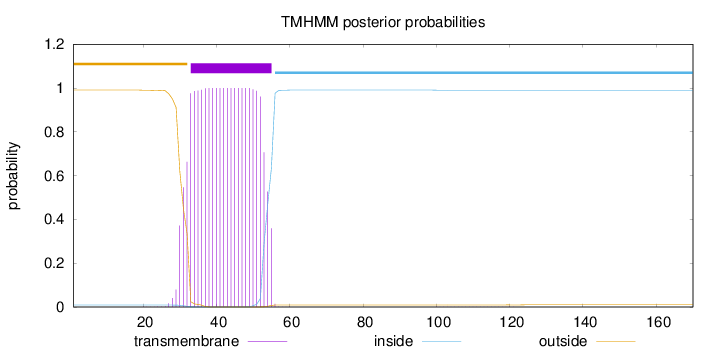
Topology
Length:
170
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.22828
Exp number, first 60 AAs:
23.22003
Total prob of N-in:
0.00934
POSSIBLE N-term signal
sequence
outside
1 - 32
TMhelix
33 - 55
inside
56 - 170
Population Genetic Test Statistics
Pi
240.988671
Theta
170.991742
Tajima's D
1.169839
CLR
0.178453
CSRT
0.710914454277286
Interpretation
Uncertain
