Gene
KWMTBOMO03991
Annotation
PREDICTED:_uncharacterized_protein_LOC106131495_isoform_X2_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.075
Sequence
CDS
ATGGAGTTAGCTAAAGTGTGTTATGTCTGGTGTCTATTGTCATTTCTGCTGTTCCAATGTCTATTGGAAGGAACTGGTCAGCAGACATCGTTCGAACTACCAGTACAGTTGATAGGTTTCCCTTTCATCATCATGGCGGTGCGACTCACCAACTTTGTTAAGAAACTATCCTATGCTTTGAGCCCAGAGACGCCATTCGAATTACCGGTACAATTAGTTGGATTCCCTTTTATCGTGACCAGTGTTAAGTTGACGAATTTCTTGAAGAAGCTGTCATATTCTTTAAGTCCAGCGACATACCGGTCTCGGAGTCGCCGTGAGTTGTTGCTAACGGACCCAGCCGAGCCCTTCGATGTCGTAGAAGTGGAGAAATACATAGTGGGCGAGTTCGGTGCCAAAGCTTGCGTGTTCGAACGTGTATGCGCGCACTATGCGACCAGGGCGCGAACGCAACCGCGGCCTCAACTTGACTGGCCAGATGTTTTCAGTCAATACAAGCGCTCTCCGGATCAAGCCAAAGAATATTATTTGCTGAGCGTGTTCCTAGGAGACATCGTGGGCTCACCCACATTATGCCATCAGCTTGGTAAGCGAATGAACTGCGACACCCAACAGTTCGGAATATAA
Protein
MELAKVCYVWCLLSFLLFQCLLEGTGQQTSFELPVQLIGFPFIIMAVRLTNFVKKLSYALSPETPFELPVQLVGFPFIVTSVKLTNFLKKLSYSLSPATYRSRSRRELLLTDPAEPFDVVEVEKYIVGEFGAKACVFERVCAHYATRARTQPRPQLDWPDVFSQYKRSPDQAKEYYLLSVFLGDIVGSPTLCHQLGKRMNCDTQQFGI
Summary
Uniprot
H9JL54
A0A2A4J6T5
A0A2H1VY01
A0A3S2PA30
A0A194R4E3
A0A194QD88
+ More
A0A154P8V2 A0A0C9PYY4 A0A0L7QRM7 A0A088ALJ6 A0A0M9A0D1 E2BIG1 A0A195DS72 A0A195FE23 F4X8L2 A0A195C8H7 A0A026W700 A0A1Y1LIH4 A0A3L8DV88 A0A182XZ31 A0A1J1ICI3 Q7QJL1 A0A182RB01 A0A139WPM7 A0A182X3I8 A0A182UPA2 A0A182KRL0 A0A182U4R1 A0A182I3I3 A0A1B0CHD3 A0A182MF85 A0A182P3R4 A0A1S3DGY6 A0A182QY49 E0VFX3 A0A182WJ11 A0A182IXI4 J9KAJ7 A0A182NH82 A0A182FI82 A0A2M4CZQ9 A0A0L7KYP7 Q17AF3 A0A0L0C8I6 A0A2S2PKS8 A0A1B0G5T9 A0A1B0BIP5 A0A1A9ZU12 A0A0M3QWY9 A0A1I8NZD2 B3M8Q6 A0A336KEK5 B4L043 A0A1I8MFZ4 B4LHX1 A0A3B0K0Y1 Q2LZ01 B4J0M9 B4N469 B3ND89 B4PJ39 A0A0A1XNI0 Q9VVN9 A0A1W4VSI1 B4QPE9 A0A034V8N3 B4IFQ6 A0A0K8VZH7 W8BR71 A0A151WF89 A0A195B9T0 A0A1A9V445 A0A1B0D6Q3
A0A154P8V2 A0A0C9PYY4 A0A0L7QRM7 A0A088ALJ6 A0A0M9A0D1 E2BIG1 A0A195DS72 A0A195FE23 F4X8L2 A0A195C8H7 A0A026W700 A0A1Y1LIH4 A0A3L8DV88 A0A182XZ31 A0A1J1ICI3 Q7QJL1 A0A182RB01 A0A139WPM7 A0A182X3I8 A0A182UPA2 A0A182KRL0 A0A182U4R1 A0A182I3I3 A0A1B0CHD3 A0A182MF85 A0A182P3R4 A0A1S3DGY6 A0A182QY49 E0VFX3 A0A182WJ11 A0A182IXI4 J9KAJ7 A0A182NH82 A0A182FI82 A0A2M4CZQ9 A0A0L7KYP7 Q17AF3 A0A0L0C8I6 A0A2S2PKS8 A0A1B0G5T9 A0A1B0BIP5 A0A1A9ZU12 A0A0M3QWY9 A0A1I8NZD2 B3M8Q6 A0A336KEK5 B4L043 A0A1I8MFZ4 B4LHX1 A0A3B0K0Y1 Q2LZ01 B4J0M9 B4N469 B3ND89 B4PJ39 A0A0A1XNI0 Q9VVN9 A0A1W4VSI1 B4QPE9 A0A034V8N3 B4IFQ6 A0A0K8VZH7 W8BR71 A0A151WF89 A0A195B9T0 A0A1A9V445 A0A1B0D6Q3
Pubmed
19121390
26354079
20798317
21719571
24508170
28004739
+ More
30249741 25244985 12364791 14747013 17210077 18362917 19820115 20966253 20566863 26227816 17510324 26108605 17994087 25315136 15632085 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 24495485
30249741 25244985 12364791 14747013 17210077 18362917 19820115 20966253 20566863 26227816 17510324 26108605 17994087 25315136 15632085 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 24495485
EMBL
BABH01011612
NWSH01002633
PCG67795.1
ODYU01005129
SOQ45693.1
RSAL01000155
+ More
RVE45646.1 KQ460930 KPJ10726.1 KQ459167 KPJ03508.1 KQ434827 KZC07648.1 GBYB01006718 JAG76485.1 KQ414775 KOC61295.1 KQ435794 KOX73766.1 GL448504 EFN84549.1 KQ980581 KYN15369.1 KQ981673 KYN38274.1 GL888932 EGI57281.1 KQ978219 KYM96428.1 KK107372 EZA51758.1 GEZM01056392 JAV72701.1 QOIP01000004 RLU24226.1 CVRI01000047 CRK97907.1 AAAB01008807 EAA04585.3 KQ973148 KYB29920.1 APCN01000213 AJWK01012240 AJWK01012241 AJWK01012242 AXCM01013404 AXCN02000893 DS235127 EEB12279.1 ABLF02038708 GGFL01006591 MBW70769.1 JTDY01004269 KOB68357.1 CH477336 EAT43207.1 JRES01000760 KNC28591.1 GGMR01017375 MBY29994.1 CCAG010010747 JXJN01015034 JXJN01015035 JXJN01015036 CP012525 ALC45013.1 CH902618 EDV41057.1 UFQS01000322 UFQT01000322 SSX02871.1 SSX23238.1 CH933809 EDW19078.1 CH940647 EDW70696.1 OUUW01000002 SPP77028.1 CH379069 EAL29708.1 CH916366 EDV97884.1 CH964095 EDW78943.1 CH954178 EDV51882.1 CM000159 EDW94630.1 GBXI01016827 GBXI01001780 JAC97464.1 JAD12512.1 AE014296 AY071344 AY113426 AAF49272.1 AAL48966.1 AAM29431.1 CM000363 CM002912 EDX10943.1 KMZ00376.1 GAKP01020842 GAKP01020840 JAC38110.1 CH480834 EDW46493.1 GDHF01008073 JAI44241.1 GAMC01010824 JAB95731.1 KQ983227 KYQ46486.1 KQ976540 KYM81286.1 AJVK01012112
RVE45646.1 KQ460930 KPJ10726.1 KQ459167 KPJ03508.1 KQ434827 KZC07648.1 GBYB01006718 JAG76485.1 KQ414775 KOC61295.1 KQ435794 KOX73766.1 GL448504 EFN84549.1 KQ980581 KYN15369.1 KQ981673 KYN38274.1 GL888932 EGI57281.1 KQ978219 KYM96428.1 KK107372 EZA51758.1 GEZM01056392 JAV72701.1 QOIP01000004 RLU24226.1 CVRI01000047 CRK97907.1 AAAB01008807 EAA04585.3 KQ973148 KYB29920.1 APCN01000213 AJWK01012240 AJWK01012241 AJWK01012242 AXCM01013404 AXCN02000893 DS235127 EEB12279.1 ABLF02038708 GGFL01006591 MBW70769.1 JTDY01004269 KOB68357.1 CH477336 EAT43207.1 JRES01000760 KNC28591.1 GGMR01017375 MBY29994.1 CCAG010010747 JXJN01015034 JXJN01015035 JXJN01015036 CP012525 ALC45013.1 CH902618 EDV41057.1 UFQS01000322 UFQT01000322 SSX02871.1 SSX23238.1 CH933809 EDW19078.1 CH940647 EDW70696.1 OUUW01000002 SPP77028.1 CH379069 EAL29708.1 CH916366 EDV97884.1 CH964095 EDW78943.1 CH954178 EDV51882.1 CM000159 EDW94630.1 GBXI01016827 GBXI01001780 JAC97464.1 JAD12512.1 AE014296 AY071344 AY113426 AAF49272.1 AAL48966.1 AAM29431.1 CM000363 CM002912 EDX10943.1 KMZ00376.1 GAKP01020842 GAKP01020840 JAC38110.1 CH480834 EDW46493.1 GDHF01008073 JAI44241.1 GAMC01010824 JAB95731.1 KQ983227 KYQ46486.1 KQ976540 KYM81286.1 AJVK01012112
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000076502
+ More
UP000053825 UP000005203 UP000053105 UP000008237 UP000078492 UP000078541 UP000007755 UP000078542 UP000053097 UP000279307 UP000076408 UP000183832 UP000007062 UP000075900 UP000007266 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000092461 UP000075883 UP000075885 UP000079169 UP000075886 UP000009046 UP000075920 UP000075880 UP000007819 UP000075884 UP000069272 UP000037510 UP000008820 UP000037069 UP000092444 UP000092460 UP000092445 UP000092553 UP000095300 UP000007801 UP000009192 UP000095301 UP000008792 UP000268350 UP000001819 UP000001070 UP000007798 UP000008711 UP000002282 UP000000803 UP000192221 UP000000304 UP000001292 UP000075809 UP000078540 UP000078200 UP000092462
UP000053825 UP000005203 UP000053105 UP000008237 UP000078492 UP000078541 UP000007755 UP000078542 UP000053097 UP000279307 UP000076408 UP000183832 UP000007062 UP000075900 UP000007266 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000092461 UP000075883 UP000075885 UP000079169 UP000075886 UP000009046 UP000075920 UP000075880 UP000007819 UP000075884 UP000069272 UP000037510 UP000008820 UP000037069 UP000092444 UP000092460 UP000092445 UP000092553 UP000095300 UP000007801 UP000009192 UP000095301 UP000008792 UP000268350 UP000001819 UP000001070 UP000007798 UP000008711 UP000002282 UP000000803 UP000192221 UP000000304 UP000001292 UP000075809 UP000078540 UP000078200 UP000092462
Pfam
PF00089 Trypsin
SUPFAM
SSF50494
SSF50494
ProteinModelPortal
H9JL54
A0A2A4J6T5
A0A2H1VY01
A0A3S2PA30
A0A194R4E3
A0A194QD88
+ More
A0A154P8V2 A0A0C9PYY4 A0A0L7QRM7 A0A088ALJ6 A0A0M9A0D1 E2BIG1 A0A195DS72 A0A195FE23 F4X8L2 A0A195C8H7 A0A026W700 A0A1Y1LIH4 A0A3L8DV88 A0A182XZ31 A0A1J1ICI3 Q7QJL1 A0A182RB01 A0A139WPM7 A0A182X3I8 A0A182UPA2 A0A182KRL0 A0A182U4R1 A0A182I3I3 A0A1B0CHD3 A0A182MF85 A0A182P3R4 A0A1S3DGY6 A0A182QY49 E0VFX3 A0A182WJ11 A0A182IXI4 J9KAJ7 A0A182NH82 A0A182FI82 A0A2M4CZQ9 A0A0L7KYP7 Q17AF3 A0A0L0C8I6 A0A2S2PKS8 A0A1B0G5T9 A0A1B0BIP5 A0A1A9ZU12 A0A0M3QWY9 A0A1I8NZD2 B3M8Q6 A0A336KEK5 B4L043 A0A1I8MFZ4 B4LHX1 A0A3B0K0Y1 Q2LZ01 B4J0M9 B4N469 B3ND89 B4PJ39 A0A0A1XNI0 Q9VVN9 A0A1W4VSI1 B4QPE9 A0A034V8N3 B4IFQ6 A0A0K8VZH7 W8BR71 A0A151WF89 A0A195B9T0 A0A1A9V445 A0A1B0D6Q3
A0A154P8V2 A0A0C9PYY4 A0A0L7QRM7 A0A088ALJ6 A0A0M9A0D1 E2BIG1 A0A195DS72 A0A195FE23 F4X8L2 A0A195C8H7 A0A026W700 A0A1Y1LIH4 A0A3L8DV88 A0A182XZ31 A0A1J1ICI3 Q7QJL1 A0A182RB01 A0A139WPM7 A0A182X3I8 A0A182UPA2 A0A182KRL0 A0A182U4R1 A0A182I3I3 A0A1B0CHD3 A0A182MF85 A0A182P3R4 A0A1S3DGY6 A0A182QY49 E0VFX3 A0A182WJ11 A0A182IXI4 J9KAJ7 A0A182NH82 A0A182FI82 A0A2M4CZQ9 A0A0L7KYP7 Q17AF3 A0A0L0C8I6 A0A2S2PKS8 A0A1B0G5T9 A0A1B0BIP5 A0A1A9ZU12 A0A0M3QWY9 A0A1I8NZD2 B3M8Q6 A0A336KEK5 B4L043 A0A1I8MFZ4 B4LHX1 A0A3B0K0Y1 Q2LZ01 B4J0M9 B4N469 B3ND89 B4PJ39 A0A0A1XNI0 Q9VVN9 A0A1W4VSI1 B4QPE9 A0A034V8N3 B4IFQ6 A0A0K8VZH7 W8BR71 A0A151WF89 A0A195B9T0 A0A1A9V445 A0A1B0D6Q3
Ontologies
Topology
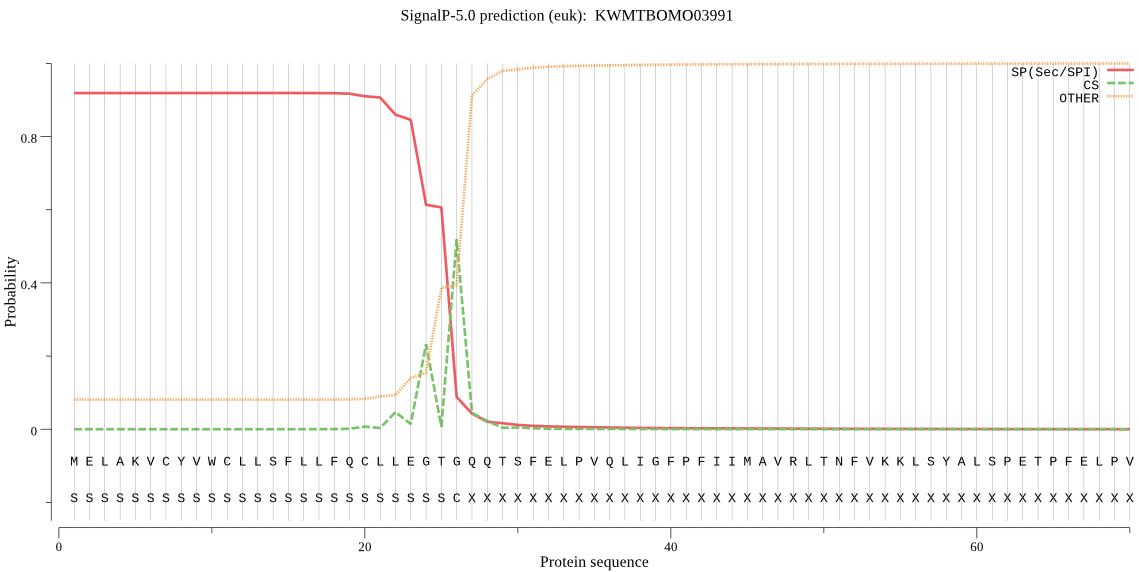
SignalP
Position: 1 - 26,
Likelihood: 0.918605
Length:
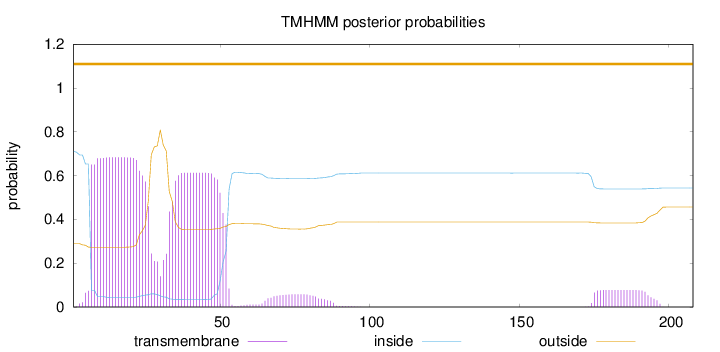
208
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
28.73747
Exp number, first 60 AAs:
25.99386
Total prob of N-in:
0.71055
POSSIBLE N-term signal
sequence
outside
1 - 208
Population Genetic Test Statistics
Pi
257.383421
Theta
172.206922
Tajima's D
1.377154
CLR
0.180431
CSRT
0.755362231888406
Interpretation
Uncertain
