Gene
KWMTBOMO03988
Pre Gene Modal
BGIBMGA010253
Annotation
PREDICTED:_copper_homeostasis_protein_cutC_homolog_isoform_X1_[Bombyx_mori]
Full name
Copper homeostasis protein CutC
+ More
Copper homeostasis protein cutC homolog
Copper homeostasis protein cutC homolog
Location in the cell
Cytoplasmic Reliability : 1.697 Nuclear Reliability : 1.323
Sequence
CDS
ATGCTTGAAGTTTGCATAGACAGTTTGGAATCCGCGATTAATGCTATTTACGGTGGTGCGGACGAGTTGGAAGTTTGTAGTTCACTTACTGAAGGCGGACTTACACCGAGCCCAGGGCTCGTATCTCAGATTGCGTCCGTCGTAAACAATATTAAAAATGGACGACTGATTCATAAATCGTGTTCTCAGTACAGCTATGGAAAGAAAACTCTTAAGGTTCCCAAAGTGAACGTCATGATAAGATGTCGCACTGGTTCGGATTTTTGTTACAACGATTTCGAAATGGACACAATGTTATCAGACATTGATCATTTCAAGCAGTTTGAAATAGACAGATTCGTTTTTGGCGCTTTAACAGATAGTCAGGAAATAGACGAAAAAAACTGCAAAAAGATTATTGAGAGAGCATCTCCTACGCCTGTGACATTCCATAGGGCGTTTGACGTTTGCAAAGAACCCAATGCTACCGTTACAAAAATTATAAATCTGGGATTTAATAGATTATTGACCAGTGGGCAGAGATGCACGGCAGCGGATTCTGAAGCTTTAGTATTAATCCGAACACTTTTAGAGAGTTTTTCTGATAAAATTGAAATTATGCCCGGAGCTGGTGTCAATGTCGATAATGCCAAACTATTCATAGATATGGGTTGTAAGATTTTGCATAGCTCGTGTAAAATAACAAAATATTTGAATAAAGCTGGCAATAATTTGAGGATGGGTGAAGGTGACAGTGAATTTATTTATGTTACTGATGAAAATATCGTGAAAGAAATGAAGCTCAAAATGGAAAATTAA
Protein
MLEVCIDSLESAINAIYGGADELEVCSSLTEGGLTPSPGLVSQIASVVNNIKNGRLIHKSCSQYSYGKKTLKVPKVNVMIRCRTGSDFCYNDFEMDTMLSDIDHFKQFEIDRFVFGALTDSQEIDEKNCKKIIERASPTPVTFHRAFDVCKEPNATVTKIINLGFNRLLTSGQRCTAADSEALVLIRTLLESFSDKIEIMPGAGVNVDNAKLFIDMGCKILHSSCKITKYLNKAGNNLRMGEGDSEFIYVTDENIVKEMKLKMEN
Summary
Description
Participates in the control of copper homeostasis.
Involved in copper homeostasis. Affects body morphology and length, egg laying and brood size.
Involved in copper homeostasis. Affects body morphology and length, egg laying and brood size.
Similarity
Belongs to the cytochrome P450 family.
Belongs to the CutC family.
Belongs to the CutC family.
Keywords
Complete proteome
Copper
Reference proteome
Feature
chain Copper homeostasis protein cutC homolog
Uniprot
A0A2A4JCD6
A0A3S2NNK2
A0A2W1BRJ9
A0A2H1WUY5
A0A194QZL8
A0A194QEY5
+ More
A0A212F9H5 S4P682 A0A151WET6 A0A151IUM2 E9IWQ7 A0A0J7KPL7 A0A151I0L9 F4WEA5 A0A151ILT0 A0A195FG38 A0A158N9Q7 A0A026WH50 A0A2J7PJP4 A0A2J7PJP7 A0A067RQ22 A0A2J7NKI4 A0A310SGD9 E2C1L5 E2A257 A0A154P2D2 A0A2A3EQW9 A0A2H8TUS9 A0A2S2PDH1 A0A088AF56 A0A0L7QYP5 A0A1Y1NI17 J9JXW9 A0A0C9PLD8 A0A0C9R4U5 A0A1L8DAR3 D6WZX5 A0A1B0DLU6 X1WKK2 A0A182MV95 N6SS60 A0A2E3SNZ5 A0A182XI82 A0A3B5K3T0 J3JU22 A0A1W4W4S1 A0A016TGS3 A0A182PXT5 A0A3B4B9N1 W5U5P9 A0A182GUV5 A0A0A9YLC9 A0A182LFB1 A0A2D0QGX2 A0A3B4Z561 W8BBE4 A0A182HT69 A0A261BKH0 A0A343VYS2 A0A368H4X4 H3DGG9 A0A3Q3WWU9 Q7QC16 A0A1I7TBD5 A0A3Q3BQ24 A0A182U814 A0A182FKW9 A0A3B3BA04 A0A1Z5LE21 A0A2G5UV49 A8XT44 P34630 A0A3B4VPN2 A0A182R3M4 A0A182KGW1 A0A3B4FPX8 A0A3Q2US65 A0A3P8P4B2 A0A3P9AU80 I3JST0 A0A2K6BUB9 A0A2K6AEP8 A0A0D9R078 A0A096P658 G7PDQ4 H9EMJ1 A0A3Q3FKY3 A0A0N0BKC2 A0A2K5KQN6 A0A182UMG2 A0A1B6DWR0 A0A1A8CPL2 A0A3Q0T9N8 F7I5K3 A0A3Q3JUN9 G7N0W5 A0A3P9HFB8 U3EID7
A0A212F9H5 S4P682 A0A151WET6 A0A151IUM2 E9IWQ7 A0A0J7KPL7 A0A151I0L9 F4WEA5 A0A151ILT0 A0A195FG38 A0A158N9Q7 A0A026WH50 A0A2J7PJP4 A0A2J7PJP7 A0A067RQ22 A0A2J7NKI4 A0A310SGD9 E2C1L5 E2A257 A0A154P2D2 A0A2A3EQW9 A0A2H8TUS9 A0A2S2PDH1 A0A088AF56 A0A0L7QYP5 A0A1Y1NI17 J9JXW9 A0A0C9PLD8 A0A0C9R4U5 A0A1L8DAR3 D6WZX5 A0A1B0DLU6 X1WKK2 A0A182MV95 N6SS60 A0A2E3SNZ5 A0A182XI82 A0A3B5K3T0 J3JU22 A0A1W4W4S1 A0A016TGS3 A0A182PXT5 A0A3B4B9N1 W5U5P9 A0A182GUV5 A0A0A9YLC9 A0A182LFB1 A0A2D0QGX2 A0A3B4Z561 W8BBE4 A0A182HT69 A0A261BKH0 A0A343VYS2 A0A368H4X4 H3DGG9 A0A3Q3WWU9 Q7QC16 A0A1I7TBD5 A0A3Q3BQ24 A0A182U814 A0A182FKW9 A0A3B3BA04 A0A1Z5LE21 A0A2G5UV49 A8XT44 P34630 A0A3B4VPN2 A0A182R3M4 A0A182KGW1 A0A3B4FPX8 A0A3Q2US65 A0A3P8P4B2 A0A3P9AU80 I3JST0 A0A2K6BUB9 A0A2K6AEP8 A0A0D9R078 A0A096P658 G7PDQ4 H9EMJ1 A0A3Q3FKY3 A0A0N0BKC2 A0A2K5KQN6 A0A182UMG2 A0A1B6DWR0 A0A1A8CPL2 A0A3Q0T9N8 F7I5K3 A0A3Q3JUN9 G7N0W5 A0A3P9HFB8 U3EID7
Pubmed
28756777
26354079
22118469
23622113
21282665
21719571
+ More
21347285 24508170 30249741 24845553 20798317 28004739 18362917 19820115 23537049 29337314 21551351 22516182 25730766 25463417 23127152 26483478 25401762 26823975 20966253 24495485 15496914 12364791 14747013 17210077 29451363 28528879 14624247 7906398 9851916 18723824 25186727 22002653 25319552 17554307 25243066
21347285 24508170 30249741 24845553 20798317 28004739 18362917 19820115 23537049 29337314 21551351 22516182 25730766 25463417 23127152 26483478 25401762 26823975 20966253 24495485 15496914 12364791 14747013 17210077 29451363 28528879 14624247 7906398 9851916 18723824 25186727 22002653 25319552 17554307 25243066
EMBL
NWSH01002120
PCG69100.1
RSAL01000202
RVE44399.1
KZ149940
PZC76941.1
+ More
ODYU01011249 SOQ56870.1 KQ460930 KPJ10729.1 KQ459167 KPJ03510.1 AGBW02009594 OWR50359.1 GAIX01006956 JAA85604.1 KQ983238 KYQ46327.1 KQ980956 KYN11097.1 GL766597 EFZ15002.1 LBMM01004560 KMQ92292.1 KQ976616 KYM79265.1 GL888102 EGI67543.1 KQ977104 KYN05823.1 KQ981606 KYN39655.1 ADTU01000141 ADTU01000142 KK107209 QOIP01000002 EZA55360.1 RLU25992.1 NEVH01024946 PNF16553.1 PNF16562.1 KK852498 KDR22695.1 NEVH01052798 PNE09479.1 KQ767877 OAD53259.1 GL451937 EFN78191.1 GL435922 EFN72462.1 KQ434803 KZC06017.1 KZ288199 PBC33616.1 GFXV01005894 MBW17699.1 GGMR01014878 MBY27497.1 KQ414688 KOC63686.1 GEZM01003861 JAV96520.1 ABLF02038519 GBYB01001860 GBYB01001863 GBYB01001864 JAG71627.1 JAG71630.1 JAG71631.1 GBYB01001861 JAG71628.1 GFDF01010562 JAV03522.1 KQ971372 EFA10815.1 AJVK01036681 ABLF02020380 ABLF02020385 ABLF02038517 ABLF02053191 AXCM01000752 APGK01059305 KB741292 KB632203 ENN70484.1 ERL90077.1 PARP01000029 MBC35193.1 BT126734 AEE61696.1 JARK01001440 EYC01860.1 JT405540 AHH37223.1 JXUM01089766 KQ563796 KXJ73289.1 GBHO01009733 GBHO01009732 GDHC01020613 GDHC01017055 JAG33871.1 JAG33872.1 JAP98015.1 JAQ01574.1 GAMC01019381 JAB87174.1 APCN01000420 NIPN01000127 OZG10240.1 KY660276 AVQ09310.1 JOJR01000012 RCN51646.1 AAAB01008859 EAA08094.4 GFJQ02001314 JAW05656.1 PDUG01000003 PIC43096.1 HE600963 CAP35648.1 FO081668 AERX01006322 AQIB01042674 AQIB01042675 AHZZ02035893 AHZZ02035894 AHZZ02035895 AQIA01072244 AQIA01072245 CM001284 EHH64936.1 JU319844 JU471797 AFE63600.1 AFH28601.1 KQ435706 KOX80298.1 GEDC01007193 JAS30105.1 HADZ01017763 SBP81704.1 CM001261 EHH19290.1 GAMT01009753 GAMS01001436 GAMR01006260 GAMQ01001535 GAMP01004738 JAB02108.1 JAB21700.1 JAB27672.1 JAB40316.1 JAB48017.1
ODYU01011249 SOQ56870.1 KQ460930 KPJ10729.1 KQ459167 KPJ03510.1 AGBW02009594 OWR50359.1 GAIX01006956 JAA85604.1 KQ983238 KYQ46327.1 KQ980956 KYN11097.1 GL766597 EFZ15002.1 LBMM01004560 KMQ92292.1 KQ976616 KYM79265.1 GL888102 EGI67543.1 KQ977104 KYN05823.1 KQ981606 KYN39655.1 ADTU01000141 ADTU01000142 KK107209 QOIP01000002 EZA55360.1 RLU25992.1 NEVH01024946 PNF16553.1 PNF16562.1 KK852498 KDR22695.1 NEVH01052798 PNE09479.1 KQ767877 OAD53259.1 GL451937 EFN78191.1 GL435922 EFN72462.1 KQ434803 KZC06017.1 KZ288199 PBC33616.1 GFXV01005894 MBW17699.1 GGMR01014878 MBY27497.1 KQ414688 KOC63686.1 GEZM01003861 JAV96520.1 ABLF02038519 GBYB01001860 GBYB01001863 GBYB01001864 JAG71627.1 JAG71630.1 JAG71631.1 GBYB01001861 JAG71628.1 GFDF01010562 JAV03522.1 KQ971372 EFA10815.1 AJVK01036681 ABLF02020380 ABLF02020385 ABLF02038517 ABLF02053191 AXCM01000752 APGK01059305 KB741292 KB632203 ENN70484.1 ERL90077.1 PARP01000029 MBC35193.1 BT126734 AEE61696.1 JARK01001440 EYC01860.1 JT405540 AHH37223.1 JXUM01089766 KQ563796 KXJ73289.1 GBHO01009733 GBHO01009732 GDHC01020613 GDHC01017055 JAG33871.1 JAG33872.1 JAP98015.1 JAQ01574.1 GAMC01019381 JAB87174.1 APCN01000420 NIPN01000127 OZG10240.1 KY660276 AVQ09310.1 JOJR01000012 RCN51646.1 AAAB01008859 EAA08094.4 GFJQ02001314 JAW05656.1 PDUG01000003 PIC43096.1 HE600963 CAP35648.1 FO081668 AERX01006322 AQIB01042674 AQIB01042675 AHZZ02035893 AHZZ02035894 AHZZ02035895 AQIA01072244 AQIA01072245 CM001284 EHH64936.1 JU319844 JU471797 AFE63600.1 AFH28601.1 KQ435706 KOX80298.1 GEDC01007193 JAS30105.1 HADZ01017763 SBP81704.1 CM001261 EHH19290.1 GAMT01009753 GAMS01001436 GAMR01006260 GAMQ01001535 GAMP01004738 JAB02108.1 JAB21700.1 JAB27672.1 JAB40316.1 JAB48017.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
UP000075809
+ More
UP000078492 UP000036403 UP000078540 UP000007755 UP000078542 UP000078541 UP000005205 UP000053097 UP000279307 UP000235965 UP000027135 UP000008237 UP000000311 UP000076502 UP000242457 UP000005203 UP000053825 UP000007819 UP000007266 UP000092462 UP000075883 UP000019118 UP000030742 UP000076407 UP000005226 UP000192223 UP000024635 UP000075885 UP000261520 UP000221080 UP000069940 UP000249989 UP000075882 UP000261400 UP000075840 UP000216463 UP000252519 UP000007303 UP000261620 UP000007062 UP000095282 UP000264800 UP000075902 UP000069272 UP000261560 UP000230233 UP000008549 UP000001940 UP000261420 UP000075900 UP000075881 UP000261460 UP000264840 UP000265100 UP000265160 UP000005207 UP000233120 UP000233140 UP000029965 UP000028761 UP000009130 UP000233100 UP000261660 UP000053105 UP000233060 UP000075903 UP000261340 UP000008225 UP000261600 UP000265200
UP000078492 UP000036403 UP000078540 UP000007755 UP000078542 UP000078541 UP000005205 UP000053097 UP000279307 UP000235965 UP000027135 UP000008237 UP000000311 UP000076502 UP000242457 UP000005203 UP000053825 UP000007819 UP000007266 UP000092462 UP000075883 UP000019118 UP000030742 UP000076407 UP000005226 UP000192223 UP000024635 UP000075885 UP000261520 UP000221080 UP000069940 UP000249989 UP000075882 UP000261400 UP000075840 UP000216463 UP000252519 UP000007303 UP000261620 UP000007062 UP000095282 UP000264800 UP000075902 UP000069272 UP000261560 UP000230233 UP000008549 UP000001940 UP000261420 UP000075900 UP000075881 UP000261460 UP000264840 UP000265100 UP000265160 UP000005207 UP000233120 UP000233140 UP000029965 UP000028761 UP000009130 UP000233100 UP000261660 UP000053105 UP000233060 UP000075903 UP000261340 UP000008225 UP000261600 UP000265200
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JCD6
A0A3S2NNK2
A0A2W1BRJ9
A0A2H1WUY5
A0A194QZL8
A0A194QEY5
+ More
A0A212F9H5 S4P682 A0A151WET6 A0A151IUM2 E9IWQ7 A0A0J7KPL7 A0A151I0L9 F4WEA5 A0A151ILT0 A0A195FG38 A0A158N9Q7 A0A026WH50 A0A2J7PJP4 A0A2J7PJP7 A0A067RQ22 A0A2J7NKI4 A0A310SGD9 E2C1L5 E2A257 A0A154P2D2 A0A2A3EQW9 A0A2H8TUS9 A0A2S2PDH1 A0A088AF56 A0A0L7QYP5 A0A1Y1NI17 J9JXW9 A0A0C9PLD8 A0A0C9R4U5 A0A1L8DAR3 D6WZX5 A0A1B0DLU6 X1WKK2 A0A182MV95 N6SS60 A0A2E3SNZ5 A0A182XI82 A0A3B5K3T0 J3JU22 A0A1W4W4S1 A0A016TGS3 A0A182PXT5 A0A3B4B9N1 W5U5P9 A0A182GUV5 A0A0A9YLC9 A0A182LFB1 A0A2D0QGX2 A0A3B4Z561 W8BBE4 A0A182HT69 A0A261BKH0 A0A343VYS2 A0A368H4X4 H3DGG9 A0A3Q3WWU9 Q7QC16 A0A1I7TBD5 A0A3Q3BQ24 A0A182U814 A0A182FKW9 A0A3B3BA04 A0A1Z5LE21 A0A2G5UV49 A8XT44 P34630 A0A3B4VPN2 A0A182R3M4 A0A182KGW1 A0A3B4FPX8 A0A3Q2US65 A0A3P8P4B2 A0A3P9AU80 I3JST0 A0A2K6BUB9 A0A2K6AEP8 A0A0D9R078 A0A096P658 G7PDQ4 H9EMJ1 A0A3Q3FKY3 A0A0N0BKC2 A0A2K5KQN6 A0A182UMG2 A0A1B6DWR0 A0A1A8CPL2 A0A3Q0T9N8 F7I5K3 A0A3Q3JUN9 G7N0W5 A0A3P9HFB8 U3EID7
A0A212F9H5 S4P682 A0A151WET6 A0A151IUM2 E9IWQ7 A0A0J7KPL7 A0A151I0L9 F4WEA5 A0A151ILT0 A0A195FG38 A0A158N9Q7 A0A026WH50 A0A2J7PJP4 A0A2J7PJP7 A0A067RQ22 A0A2J7NKI4 A0A310SGD9 E2C1L5 E2A257 A0A154P2D2 A0A2A3EQW9 A0A2H8TUS9 A0A2S2PDH1 A0A088AF56 A0A0L7QYP5 A0A1Y1NI17 J9JXW9 A0A0C9PLD8 A0A0C9R4U5 A0A1L8DAR3 D6WZX5 A0A1B0DLU6 X1WKK2 A0A182MV95 N6SS60 A0A2E3SNZ5 A0A182XI82 A0A3B5K3T0 J3JU22 A0A1W4W4S1 A0A016TGS3 A0A182PXT5 A0A3B4B9N1 W5U5P9 A0A182GUV5 A0A0A9YLC9 A0A182LFB1 A0A2D0QGX2 A0A3B4Z561 W8BBE4 A0A182HT69 A0A261BKH0 A0A343VYS2 A0A368H4X4 H3DGG9 A0A3Q3WWU9 Q7QC16 A0A1I7TBD5 A0A3Q3BQ24 A0A182U814 A0A182FKW9 A0A3B3BA04 A0A1Z5LE21 A0A2G5UV49 A8XT44 P34630 A0A3B4VPN2 A0A182R3M4 A0A182KGW1 A0A3B4FPX8 A0A3Q2US65 A0A3P8P4B2 A0A3P9AU80 I3JST0 A0A2K6BUB9 A0A2K6AEP8 A0A0D9R078 A0A096P658 G7PDQ4 H9EMJ1 A0A3Q3FKY3 A0A0N0BKC2 A0A2K5KQN6 A0A182UMG2 A0A1B6DWR0 A0A1A8CPL2 A0A3Q0T9N8 F7I5K3 A0A3Q3JUN9 G7N0W5 A0A3P9HFB8 U3EID7
PDB
3IWP
E-value=2.55229e-38,
Score=396
Ontologies
GO
GO:0055070
GO:0005507
GO:0005506
GO:0016705
GO:0020037
GO:0004497
GO:0004037
GO:0000256
GO:0006878
GO:0005737
GO:0040025
GO:0046688
GO:0035264
GO:0018991
GO:0030054
GO:0032504
GO:0005829
GO:0005634
GO:0055120
GO:0051262
GO:0005654
GO:0005730
GO:0006281
GO:0008094
GO:0005216
GO:0006811
GO:0006032
GO:0016021
GO:0004672
GO:0048384
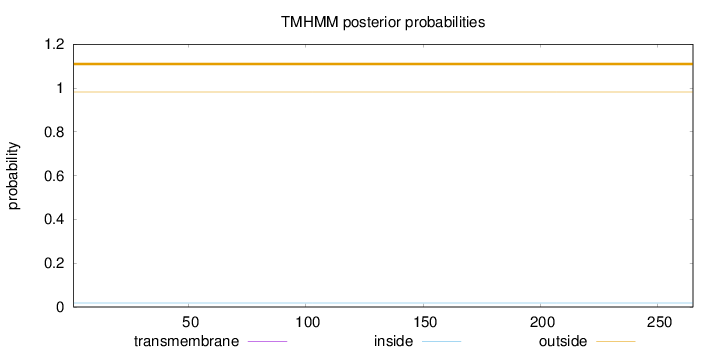
Topology
Subcellular location
Cytoplasm
Length:
265
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00161
Exp number, first 60 AAs:
0.00138
Total prob of N-in:
0.01835
outside
1 - 265
Population Genetic Test Statistics
Pi
255.128728
Theta
160.354116
Tajima's D
1.596869
CLR
0.007939
CSRT
0.809409529523524
Interpretation
Uncertain
