Gene
KWMTBOMO03986 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010130
Annotation
Annexin_IX_isoform_B_[Bombyx_mori]
Full name
Annexin
+ More
Annexin B9
Annexin B9
Alternative Name
Annexin IX
Annexin-9
Annexin-9
Location in the cell
Cytoplasmic Reliability : 3.496
Sequence
CDS
ATGAGCGGACAACAATACTACCCTTACAAGTGCACCCCCACGGTGTACCCCGCGGAGCCGTTCGACCCGGCCGCGGATGCCGAGACCCTTCGCAAGGCTATGAAGGGCTTCGGCACCGATGAGAAGGCTATCATCGACGTGCTCTGCCGCCGCGGCATCGTGCAACGCCTCGAGATCGCGGAGACCTTCAAGACTAACTATGGCAAGGACTTGATTAGCGAACTCAAGAGTGAACTCACCGGCAACTTGGAAAATGTCATCGTCGCATTGATGACTCCCCTGCCCCACTTCTACGCTAAGGAGCTCCACGATGCTGTCTCAGGAATTGGAACCGACGAAGAAGCCATCATCGAGATCCTGTGCACGCTTTCCAACTATGGTATCCGTACCATATCCGCATTTTACGAACAACTGTACGGCAAGAGCCTGGAATCGGACTTAAAAGGCGACACGTCGGGACACTTCAAGAGATTGTGCGTGTCGTTGTGCATGGCCAATCGCGATGAAAACCAGGGCATCGATGAAGGCTCAGCTAAAGCCGATGCTGAAGCACTGGCCGCCGCTGGTGAAGGTCAATGGGGAACCGACGAATCAATCTTCAACTCCATCCTTATCACTCGCTCCTATCAGCAGCTGAGACAGATCTTCGCCGAGTACGAAGCTTTGACCGGAAAAGACATTGAGGACTCGATCAAGAAAGAATTCTCTGGTAGCATTGAGAAGGGCATGCTCGCTATTGCCAAATGCGTAAAGAGCAAGGTCGGTTTCTTCGCTGAGCGCCTGTACTACTCGATGAAGGGCATCGGCACCAACGACAAGACGCTCATCCGCATCGTGGTGAGCCGCTCCGAGATCGACCTAGGGGACATCAAGCAGGCATTCCTCGAGAAGTACGGCAAGTCCCTGGAGACCTGGATCGCTGACGAAATCGCTGGTCCCCTCGGCGACCTCCTCTCGACAATGTGTTACTAG
Protein
MSGQQYYPYKCTPTVYPAEPFDPAADAETLRKAMKGFGTDEKAIIDVLCRRGIVQRLEIAETFKTNYGKDLISELKSELTGNLENVIVALMTPLPHFYAKELHDAVSGIGTDEEAIIEILCTLSNYGIRTISAFYEQLYGKSLESDLKGDTSGHFKRLCVSLCMANRDENQGIDEGSAKADAEALAAAGEGQWGTDESIFNSILITRSYQQLRQIFAEYEALTGKDIEDSIKKEFSGSIEKGMLAIAKCVKSKVGFFAERLYYSMKGIGTNDKTLIRIVVSRSEIDLGDIKQAFLEKYGKSLETWIADEIAGPLGDLLSTMCY
Summary
Similarity
Belongs to the annexin family.
Keywords
Alternative splicing
Annexin
Calcium
Complete proteome
Reference proteome
Repeat
Feature
chain Annexin B9
splice variant In isoform b.
splice variant In isoform b.
Uniprot
Q9NL60
Q9GNG6
I4DJ02
Q5F320
A0A1E1WHP3
A0A2A4JC22
+ More
H9JKS9 Q9NL59 Q9NL61 A0A1E1WTN1 Q5F321 A0A194QDH7 A0A2A4JAJ3 A0A1E1WK32 A0A1E1WKW0 A0A3S2L2U7 A0A3G1T1A2 A0A2H1W664 F5BYI5 A0A2A4JBG3 A0A2W1BPH9 A0A2A4JCI0 A0A2A4JAG6 A0A2A4JBN3 S4PS71 A0A194R4E8 V5GS56 A0A0L7LJF6 A0A3L8DCU3 V5GW36 E2BIN4 A0A026WCV9 V9IBH3 K7IYR5 J3JW49 V9ID93 A0A158P217 E2ACR5 E9J8L4 F4W699 A0A0L7R3I5 A0A0T6BE53 A0A2J7PIB0 A0A232ETP2 A0A1I9WLG9 D6WDY4 U5EV42 A0A1D8DD02 A0A0K8VHY0 A0A0C9RIT6 A0A034VS69 A0A0C9RSC7 A0A0A1X5M1 A0A1L8EEC4 A0A1L8EES1 A0A0R3NR84 A0A067RHR9 A0A1B0A555 A0A1Y1LSX9 A0A0R3NKI2 W8BK47 A0A1I8Q7T7 A0A034VRB9 D3TRS6 A0A2M4CSW8 A0A0A1WCS5 A0A0P8YFX1 Q16QE9 P22464-2 A0A1I8MR12 T1PKP5 A0A182IT98 A0A2A3EA07 A0A1Y1LS11 A0A1I8MR20 A0A023ENJ7 A0A1W4VYM2 A0A3B0KEC6 B4GM07 Q293K1 P22464 A0A2Y9D182 Q5TVB3 A0A0P9A0N5 A0A3F2YW62 A0A1I8Q7Q1 A0A1Y9IW81 A0A2Y9D2T8 A0A2Y9D0P5 A0A182FSD6 A0A1W4VXR5 A0A1Q3FIP8 B4JSX5 A0A0K8VW14 A0A0C9RSH3 A0A034VSX7 Q1HR06 A0A0Q9WVB4 A0A3B0JSK3 P22464-3
H9JKS9 Q9NL59 Q9NL61 A0A1E1WTN1 Q5F321 A0A194QDH7 A0A2A4JAJ3 A0A1E1WK32 A0A1E1WKW0 A0A3S2L2U7 A0A3G1T1A2 A0A2H1W664 F5BYI5 A0A2A4JBG3 A0A2W1BPH9 A0A2A4JCI0 A0A2A4JAG6 A0A2A4JBN3 S4PS71 A0A194R4E8 V5GS56 A0A0L7LJF6 A0A3L8DCU3 V5GW36 E2BIN4 A0A026WCV9 V9IBH3 K7IYR5 J3JW49 V9ID93 A0A158P217 E2ACR5 E9J8L4 F4W699 A0A0L7R3I5 A0A0T6BE53 A0A2J7PIB0 A0A232ETP2 A0A1I9WLG9 D6WDY4 U5EV42 A0A1D8DD02 A0A0K8VHY0 A0A0C9RIT6 A0A034VS69 A0A0C9RSC7 A0A0A1X5M1 A0A1L8EEC4 A0A1L8EES1 A0A0R3NR84 A0A067RHR9 A0A1B0A555 A0A1Y1LSX9 A0A0R3NKI2 W8BK47 A0A1I8Q7T7 A0A034VRB9 D3TRS6 A0A2M4CSW8 A0A0A1WCS5 A0A0P8YFX1 Q16QE9 P22464-2 A0A1I8MR12 T1PKP5 A0A182IT98 A0A2A3EA07 A0A1Y1LS11 A0A1I8MR20 A0A023ENJ7 A0A1W4VYM2 A0A3B0KEC6 B4GM07 Q293K1 P22464 A0A2Y9D182 Q5TVB3 A0A0P9A0N5 A0A3F2YW62 A0A1I8Q7Q1 A0A1Y9IW81 A0A2Y9D2T8 A0A2Y9D0P5 A0A182FSD6 A0A1W4VXR5 A0A1Q3FIP8 B4JSX5 A0A0K8VW14 A0A0C9RSH3 A0A034VSX7 Q1HR06 A0A0Q9WVB4 A0A3B0JSK3 P22464-3
Pubmed
11719064
11222941
22651552
19121390
26354079
21452889
+ More
28756777 23622113 26227816 30249741 20798317 24508170 20075255 22516182 23537049 21347285 21282665 21719571 28648823 27538518 18362917 19820115 27142481 25348373 25830018 15632085 17994087 23185243 24845553 28004739 24495485 20353571 17510324 10731132 12537572 12537569 2141610 25315136 24945155 20966253 12364791 14747013 17210077 18057021 17204158
28756777 23622113 26227816 30249741 20798317 24508170 20075255 22516182 23537049 21347285 21282665 21719571 28648823 27538518 18362917 19820115 27142481 25348373 25830018 15632085 17994087 23185243 24845553 28004739 24495485 20353571 17510324 10731132 12537572 12537569 2141610 25315136 24945155 20966253 12364791 14747013 17210077 18057021 17204158
EMBL
AB030497
BAA92810.1
AB041636
AB041637
BAB16697.1
BAB16698.1
+ More
AK401270 BAM17892.1 AJ863122 CAI06089.1 GDQN01004539 JAT86515.1 NWSH01002120 PCG69104.1 BABH01011600 AB030498 BAA92811.1 AB030496 BAA92809.1 GDQN01000858 JAT90196.1 AJ863121 CAI06088.1 KQ459167 KPJ03512.1 PCG69105.1 GDQN01006430 GDQN01003691 JAT84624.1 JAT87363.1 GDQN01003548 JAT87506.1 RSAL01000202 RVE44401.1 MG846909 AXY94761.1 ODYU01006521 SOQ48446.1 JF417990 AEB26321.1 PCG69106.1 KZ149940 PZC76942.1 PCG69103.1 PCG69107.1 PCG69108.1 GAIX01014098 JAA78462.1 KQ460930 KPJ10731.1 GALX01004039 JAB64427.1 JTDY01000877 KOB75587.1 QOIP01000010 RLU17709.1 GALX01004038 JAB64428.1 GL448516 EFN84481.1 KK107262 EZA53910.1 JR038908 AEY58450.1 APGK01030351 APGK01030352 BT127467 KB740770 KB632132 AEE62429.1 ENN78998.1 ERL89002.1 JR038909 AEY58451.1 ADTU01006961 GL438568 EFN68809.1 GL769036 EFZ10787.1 GL887707 EGI70234.1 KQ414663 KOC65442.1 LJIG01001749 KRT85191.1 NEVH01025127 PNF16086.1 NNAY01002247 OXU21730.1 KU932353 APA33989.1 KQ971324 EFA01193.1 GANO01001188 JAB58683.1 KU366704 AOS87597.1 GDHF01014707 GDHF01013805 JAI37607.1 JAI38509.1 GBYB01008165 JAG77932.1 GAKP01014302 GAKP01014299 JAC44650.1 GBYB01011555 GBYB01011556 JAG81322.1 JAG81323.1 GBXI01008252 JAD06040.1 GFDG01001749 JAV17050.1 GFDG01001746 JAV17053.1 CM000070 KRT01408.1 KK852463 KDR23347.1 GEZM01051834 JAV74676.1 KRT01410.1 GAMC01016606 JAB89949.1 GAKP01014305 JAC44647.1 EZ424128 ADD20404.1 GGFL01004266 MBW68444.1 GBXI01017610 JAC96681.1 CH902617 KPU80293.1 CH477750 EAT36627.1 AF261718 AY007377 AE014297 AY118504 BT001749 BT044420 M34068 KA649284 KA649338 AFP63913.1 KZ288322 PBC28122.1 GEZM01051835 JAV74675.1 GAPW01002611 JAC10987.1 OUUW01000007 SPP83391.1 CH479185 EDW38581.1 EAL29214.2 AAAB01008844 EAL41340.2 KPU80291.1 APCN01003119 APCN01003120 GFDL01007554 JAV27491.1 CH916373 EDV94865.1 GDHF01027303 GDHF01023701 GDHF01020279 GDHF01018912 GDHF01009243 GDHF01007741 GDHF01005963 JAI25011.1 JAI28613.1 JAI32035.1 JAI33402.1 JAI43071.1 JAI44573.1 JAI46351.1 GBYB01011595 JAG81362.1 GAKP01014304 GAKP01014303 JAC44648.1 DQ440288 ABF18321.1 EAT36628.1 CH964272 KRG00070.1 SPP83392.1
AK401270 BAM17892.1 AJ863122 CAI06089.1 GDQN01004539 JAT86515.1 NWSH01002120 PCG69104.1 BABH01011600 AB030498 BAA92811.1 AB030496 BAA92809.1 GDQN01000858 JAT90196.1 AJ863121 CAI06088.1 KQ459167 KPJ03512.1 PCG69105.1 GDQN01006430 GDQN01003691 JAT84624.1 JAT87363.1 GDQN01003548 JAT87506.1 RSAL01000202 RVE44401.1 MG846909 AXY94761.1 ODYU01006521 SOQ48446.1 JF417990 AEB26321.1 PCG69106.1 KZ149940 PZC76942.1 PCG69103.1 PCG69107.1 PCG69108.1 GAIX01014098 JAA78462.1 KQ460930 KPJ10731.1 GALX01004039 JAB64427.1 JTDY01000877 KOB75587.1 QOIP01000010 RLU17709.1 GALX01004038 JAB64428.1 GL448516 EFN84481.1 KK107262 EZA53910.1 JR038908 AEY58450.1 APGK01030351 APGK01030352 BT127467 KB740770 KB632132 AEE62429.1 ENN78998.1 ERL89002.1 JR038909 AEY58451.1 ADTU01006961 GL438568 EFN68809.1 GL769036 EFZ10787.1 GL887707 EGI70234.1 KQ414663 KOC65442.1 LJIG01001749 KRT85191.1 NEVH01025127 PNF16086.1 NNAY01002247 OXU21730.1 KU932353 APA33989.1 KQ971324 EFA01193.1 GANO01001188 JAB58683.1 KU366704 AOS87597.1 GDHF01014707 GDHF01013805 JAI37607.1 JAI38509.1 GBYB01008165 JAG77932.1 GAKP01014302 GAKP01014299 JAC44650.1 GBYB01011555 GBYB01011556 JAG81322.1 JAG81323.1 GBXI01008252 JAD06040.1 GFDG01001749 JAV17050.1 GFDG01001746 JAV17053.1 CM000070 KRT01408.1 KK852463 KDR23347.1 GEZM01051834 JAV74676.1 KRT01410.1 GAMC01016606 JAB89949.1 GAKP01014305 JAC44647.1 EZ424128 ADD20404.1 GGFL01004266 MBW68444.1 GBXI01017610 JAC96681.1 CH902617 KPU80293.1 CH477750 EAT36627.1 AF261718 AY007377 AE014297 AY118504 BT001749 BT044420 M34068 KA649284 KA649338 AFP63913.1 KZ288322 PBC28122.1 GEZM01051835 JAV74675.1 GAPW01002611 JAC10987.1 OUUW01000007 SPP83391.1 CH479185 EDW38581.1 EAL29214.2 AAAB01008844 EAL41340.2 KPU80291.1 APCN01003119 APCN01003120 GFDL01007554 JAV27491.1 CH916373 EDV94865.1 GDHF01027303 GDHF01023701 GDHF01020279 GDHF01018912 GDHF01009243 GDHF01007741 GDHF01005963 JAI25011.1 JAI28613.1 JAI32035.1 JAI33402.1 JAI43071.1 JAI44573.1 JAI46351.1 GBYB01011595 JAG81362.1 GAKP01014304 GAKP01014303 JAC44648.1 DQ440288 ABF18321.1 EAT36628.1 CH964272 KRG00070.1 SPP83392.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000283053
UP000053240
UP000037510
+ More
UP000279307 UP000008237 UP000053097 UP000002358 UP000019118 UP000030742 UP000005205 UP000000311 UP000007755 UP000053825 UP000235965 UP000215335 UP000007266 UP000001819 UP000027135 UP000092445 UP000095300 UP000007801 UP000008820 UP000000803 UP000095301 UP000075880 UP000242457 UP000192221 UP000268350 UP000008744 UP000075882 UP000007062 UP000075885 UP000076407 UP000075903 UP000075840 UP000069272 UP000001070 UP000007798
UP000279307 UP000008237 UP000053097 UP000002358 UP000019118 UP000030742 UP000005205 UP000000311 UP000007755 UP000053825 UP000235965 UP000215335 UP000007266 UP000001819 UP000027135 UP000092445 UP000095300 UP000007801 UP000008820 UP000000803 UP000095301 UP000075880 UP000242457 UP000192221 UP000268350 UP000008744 UP000075882 UP000007062 UP000075885 UP000076407 UP000075903 UP000075840 UP000069272 UP000001070 UP000007798
Pfam
PF00191 Annexin
Interpro
Gene 3D
ProteinModelPortal
Q9NL60
Q9GNG6
I4DJ02
Q5F320
A0A1E1WHP3
A0A2A4JC22
+ More
H9JKS9 Q9NL59 Q9NL61 A0A1E1WTN1 Q5F321 A0A194QDH7 A0A2A4JAJ3 A0A1E1WK32 A0A1E1WKW0 A0A3S2L2U7 A0A3G1T1A2 A0A2H1W664 F5BYI5 A0A2A4JBG3 A0A2W1BPH9 A0A2A4JCI0 A0A2A4JAG6 A0A2A4JBN3 S4PS71 A0A194R4E8 V5GS56 A0A0L7LJF6 A0A3L8DCU3 V5GW36 E2BIN4 A0A026WCV9 V9IBH3 K7IYR5 J3JW49 V9ID93 A0A158P217 E2ACR5 E9J8L4 F4W699 A0A0L7R3I5 A0A0T6BE53 A0A2J7PIB0 A0A232ETP2 A0A1I9WLG9 D6WDY4 U5EV42 A0A1D8DD02 A0A0K8VHY0 A0A0C9RIT6 A0A034VS69 A0A0C9RSC7 A0A0A1X5M1 A0A1L8EEC4 A0A1L8EES1 A0A0R3NR84 A0A067RHR9 A0A1B0A555 A0A1Y1LSX9 A0A0R3NKI2 W8BK47 A0A1I8Q7T7 A0A034VRB9 D3TRS6 A0A2M4CSW8 A0A0A1WCS5 A0A0P8YFX1 Q16QE9 P22464-2 A0A1I8MR12 T1PKP5 A0A182IT98 A0A2A3EA07 A0A1Y1LS11 A0A1I8MR20 A0A023ENJ7 A0A1W4VYM2 A0A3B0KEC6 B4GM07 Q293K1 P22464 A0A2Y9D182 Q5TVB3 A0A0P9A0N5 A0A3F2YW62 A0A1I8Q7Q1 A0A1Y9IW81 A0A2Y9D2T8 A0A2Y9D0P5 A0A182FSD6 A0A1W4VXR5 A0A1Q3FIP8 B4JSX5 A0A0K8VW14 A0A0C9RSH3 A0A034VSX7 Q1HR06 A0A0Q9WVB4 A0A3B0JSK3 P22464-3
H9JKS9 Q9NL59 Q9NL61 A0A1E1WTN1 Q5F321 A0A194QDH7 A0A2A4JAJ3 A0A1E1WK32 A0A1E1WKW0 A0A3S2L2U7 A0A3G1T1A2 A0A2H1W664 F5BYI5 A0A2A4JBG3 A0A2W1BPH9 A0A2A4JCI0 A0A2A4JAG6 A0A2A4JBN3 S4PS71 A0A194R4E8 V5GS56 A0A0L7LJF6 A0A3L8DCU3 V5GW36 E2BIN4 A0A026WCV9 V9IBH3 K7IYR5 J3JW49 V9ID93 A0A158P217 E2ACR5 E9J8L4 F4W699 A0A0L7R3I5 A0A0T6BE53 A0A2J7PIB0 A0A232ETP2 A0A1I9WLG9 D6WDY4 U5EV42 A0A1D8DD02 A0A0K8VHY0 A0A0C9RIT6 A0A034VS69 A0A0C9RSC7 A0A0A1X5M1 A0A1L8EEC4 A0A1L8EES1 A0A0R3NR84 A0A067RHR9 A0A1B0A555 A0A1Y1LSX9 A0A0R3NKI2 W8BK47 A0A1I8Q7T7 A0A034VRB9 D3TRS6 A0A2M4CSW8 A0A0A1WCS5 A0A0P8YFX1 Q16QE9 P22464-2 A0A1I8MR12 T1PKP5 A0A182IT98 A0A2A3EA07 A0A1Y1LS11 A0A1I8MR20 A0A023ENJ7 A0A1W4VYM2 A0A3B0KEC6 B4GM07 Q293K1 P22464 A0A2Y9D182 Q5TVB3 A0A0P9A0N5 A0A3F2YW62 A0A1I8Q7Q1 A0A1Y9IW81 A0A2Y9D2T8 A0A2Y9D0P5 A0A182FSD6 A0A1W4VXR5 A0A1Q3FIP8 B4JSX5 A0A0K8VW14 A0A0C9RSH3 A0A034VSX7 Q1HR06 A0A0Q9WVB4 A0A3B0JSK3 P22464-3
PDB
1AEI
E-value=2.00968e-71,
Score=683
Ontologies
GO
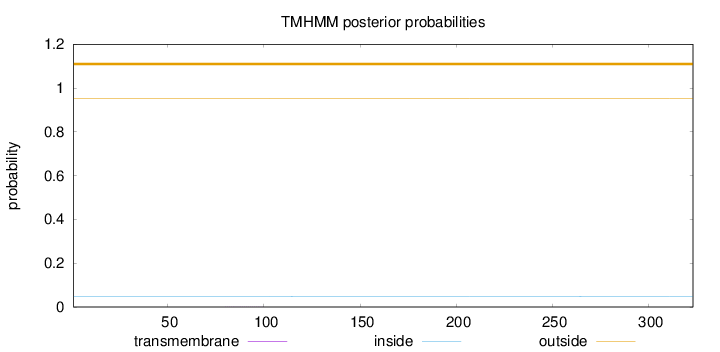
Topology
Length:
323
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00703999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04898
outside
1 - 323
Population Genetic Test Statistics
Pi
240.577212
Theta
186.335132
Tajima's D
0.890888
CLR
0.089438
CSRT
0.626868656567172
Interpretation
Possibly Positive selection
