Pre Gene Modal
BGIBMGA010248
Annotation
PREDICTED:_mitochondrial_glutamate_carrier_1-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.506 PlasmaMembrane Reliability : 1.795
Sequence
CDS
ATGAGTGCAAACATGAAACCAGTGAATCCACCTCCTCAGCAATTTTCATTAGTACCGAAGATCATCAATGGCGGTATCGCGGGTATTGTCGGGGTGTCGGTCGTGTTTCCGCTCGATCTGGTCAAGACACGCTTACAGAATCAAACAGTGGGACCCAGCGGAGAGAAGCAATATAAGAGCATGTTGGATTGCTTCAAGAAGACATACGCAGCGGAGGGCTACTTCGGAATGTATCGCGGCTCAGCCGTGAACATTATATTGATAACGCCCGAGAAAGCGATCAAACTCGCCGCAAACGACATGTTCCGACACTACCTCACTTTATCTGACGGGTCATTGCCAGTATTCCGTCAGATGGTTGCCGGTGGCTTGGCGGGAGCTTGCCAAATCGTGATAACCACGCCAATGGAACTCCTCAAAATTCAGATGCAGGACGCTGGAAGGATTGCCGCTCAAGCCAAAGCAGAGGGCCGCACGATACAGCGCACCACGGCTCTAGAGCTGACGAAGAAGTTGCTACGTGAGCGCGGTATATTCGGCCTGTACAAGGGAGTGACGGCGACGGCCGCCCGGGACGTGTCCTTCAGCGTCGTGTACTTTCCGTTGTTCGCGACCCTCAACGACCTCGGACCTAAGGAGGGGCCCGGCGGTCATACACCGTTCTGGTGGTCGTTCATATCAGGTTGTGCTGCTGGCTCCACAGCAGCGTTAGCTGTAAACCCCATGGACGTGGTGAAGACTCGCATGCAGACCCTCACCAAGGGAAGCGGAGAGAGGCAATACACCTCCATCATGGATTGTGTTACGAAAACTTTGAAATATGAAGGTCCGACTGCCTTTTTCAAAGGCGGCGCTTGCCGTATGATTGTGATCGCCCCGCTGTTCGGTATAGCTCAGGCTGTGTATTACGTTGGGATCGCCGAATATCTGCTCGGTCTCAAGTAA
Protein
MSANMKPVNPPPQQFSLVPKIINGGIAGIVGVSVVFPLDLVKTRLQNQTVGPSGEKQYKSMLDCFKKTYAAEGYFGMYRGSAVNIILITPEKAIKLAANDMFRHYLTLSDGSLPVFRQMVAGGLAGACQIVITTPMELLKIQMQDAGRIAAQAKAEGRTIQRTTALELTKKLLRERGIFGLYKGVTATAARDVSFSVVYFPLFATLNDLGPKEGPGGHTPFWWSFISGCAAGSTAALAVNPMDVVKTRMQTLTKGSGERQYTSIMDCVTKTLKYEGPTAFFKGGACRMIVIAPLFGIAQAVYYVGIAEYLLGLK
Summary
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
H9JL47
A0A3S2P8F5
A0A194QDA3
A0A2A4JG98
A0A2W1BPI9
A0A2H1WAS2
+ More
A0A1J1J183 B4GFP4 Q295T9 A0A0K8TTJ3 B3MTX1 A0A3B0JQP7 D6WN36 B4GFP5 A0A1Q3G411 A0A182N8X8 Q5TNS1 A0A182RAG4 A0A182KCQ0 A0A182VSR3 A0A0Q9WEG4 B4PN09 A0A182QU83 A0A0L7RGU2 B4K5M6 A0A310SH91 B4LWS8 A0A1W4UN02 A0A0L0BUP9 B4HHJ3 B4QTJ0 A0A023EN96 A0A336L7W8 Q9VGF7 B3P4D0 A0A1A9VPJ8 Q1HQQ1 Q16QJ1 A0A0N0U648 A0A1B6LSY0 A0A1I8P125 A0A1L8EFP8 A0A2M3Z8W5 A0A1B6KS47 A0A2M4BU31 A0A1I8N795 A0A1I8N794 W8B6Z1 W5JRA3 T1PGN7 A0A2M4AL52 A0A2A3ERV0 V9IK22 A0A1B6G8Z8 A0A158NPX9 B4JT68 U5EUW4 A0A0K8UBS0 A0A034WBY0 A0A1B6DKT8 A0A1L8E0G9 A0A195EM13 E9J6L7 A0A1A9XU33 A0A1B0AVI4 A0A1B6IM18 A0A1L8E0Y7 A0A1B0GCX7 E2B5H7 A0A182F8U5 A0A182PTW5 A0A1A9ZJQ4 A0A3L8D8X7 A0A195ATV9 A0A0M4ET21 A0A194QYX7 A0A195ESU9 A0A0M4EUR8 A0A026X2S8 A0A151X8M8 A0A0N7Z952 F4W7J9 A0A0P4VZG2 E2AT59 A0A1Y1KUJ1 A0A0A9WSU9 A0A151I6B7 T1PE37 K7IMW7 N6SW13 J9JUX6 U4UFS3 A0A224XUK3 A0A2S2QFA0 A0A232F9R8 A0A0M4EDK7 A0A0V0G6W4 A0A336LGC9 A0A084W5P6 A0A2H8TI21
A0A1J1J183 B4GFP4 Q295T9 A0A0K8TTJ3 B3MTX1 A0A3B0JQP7 D6WN36 B4GFP5 A0A1Q3G411 A0A182N8X8 Q5TNS1 A0A182RAG4 A0A182KCQ0 A0A182VSR3 A0A0Q9WEG4 B4PN09 A0A182QU83 A0A0L7RGU2 B4K5M6 A0A310SH91 B4LWS8 A0A1W4UN02 A0A0L0BUP9 B4HHJ3 B4QTJ0 A0A023EN96 A0A336L7W8 Q9VGF7 B3P4D0 A0A1A9VPJ8 Q1HQQ1 Q16QJ1 A0A0N0U648 A0A1B6LSY0 A0A1I8P125 A0A1L8EFP8 A0A2M3Z8W5 A0A1B6KS47 A0A2M4BU31 A0A1I8N795 A0A1I8N794 W8B6Z1 W5JRA3 T1PGN7 A0A2M4AL52 A0A2A3ERV0 V9IK22 A0A1B6G8Z8 A0A158NPX9 B4JT68 U5EUW4 A0A0K8UBS0 A0A034WBY0 A0A1B6DKT8 A0A1L8E0G9 A0A195EM13 E9J6L7 A0A1A9XU33 A0A1B0AVI4 A0A1B6IM18 A0A1L8E0Y7 A0A1B0GCX7 E2B5H7 A0A182F8U5 A0A182PTW5 A0A1A9ZJQ4 A0A3L8D8X7 A0A195ATV9 A0A0M4ET21 A0A194QYX7 A0A195ESU9 A0A0M4EUR8 A0A026X2S8 A0A151X8M8 A0A0N7Z952 F4W7J9 A0A0P4VZG2 E2AT59 A0A1Y1KUJ1 A0A0A9WSU9 A0A151I6B7 T1PE37 K7IMW7 N6SW13 J9JUX6 U4UFS3 A0A224XUK3 A0A2S2QFA0 A0A232F9R8 A0A0M4EDK7 A0A0V0G6W4 A0A336LGC9 A0A084W5P6 A0A2H8TI21
Pubmed
19121390
26354079
28756777
17994087
15632085
23185243
+ More
26369729 18057021 18362917 19820115 12364791 17550304 26108605 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17204158 17510324 25315136 24495485 20920257 23761445 21347285 25348373 21282665 20798317 30249741 24508170 27129103 21719571 28004739 25401762 26823975 20075255 23537049 28648823 24438588
26369729 18057021 18362917 19820115 12364791 17550304 26108605 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17204158 17510324 25315136 24495485 20920257 23761445 21347285 25348373 21282665 20798317 30249741 24508170 27129103 21719571 28004739 25401762 26823975 20075255 23537049 28648823 24438588
EMBL
BABH01011587
BABH01011588
BABH01011589
RSAL01000202
RVE44411.1
KQ459167
+ More
KPJ03523.1 NWSH01001580 PCG70806.1 KZ149940 PZC76952.1 ODYU01007415 SOQ50158.1 CVRI01000066 CRL06205.1 CH479182 EDW34429.1 CM000070 EAL28619.2 KRT00892.1 GDAI01000130 JAI17473.1 CH902623 EDV30252.1 KPU72753.1 OUUW01000008 SPP84497.1 KQ971343 EFA03243.2 EDW34430.1 GFDL01000508 JAV34537.1 AAAB01008980 EAL39319.2 CH940650 KRF82842.1 CM000160 EDW97021.1 AXCN02001876 KQ414592 KOC70197.1 CH933806 EDW14063.2 KQ761155 OAD58144.1 EDW66649.1 KRF82843.1 KRF82844.1 JRES01001304 KNC23718.1 CH480815 EDW42532.1 CM000364 EDX13295.1 GAPW01002731 JAC10867.1 UFQS01002045 UFQT01002045 SSX13061.1 SSX32501.1 AE014297 AY119214 AAF54725.1 AAM51074.1 AGB95872.1 CH954181 EDV49445.1 DQ440393 ABF18426.1 CH477747 EAT36649.1 KQ435756 KOX75978.1 GEBQ01013191 JAT26786.1 GFDG01001259 JAV17540.1 GGFM01004179 MBW24930.1 GEBQ01025720 JAT14257.1 GGFJ01007436 MBW56577.1 GAMC01013712 GAMC01013711 JAB92843.1 ADMH02000408 ETN66666.1 KA646388 KA647028 AFP61657.1 GGFK01008190 MBW41511.1 KZ288191 PBC34420.1 JR049541 AEY60997.1 GECZ01024041 GECZ01010888 GECZ01010132 JAS45728.1 JAS58881.1 JAS59637.1 ADTU01022691 CH916373 EDV94958.1 GANO01002145 JAB57726.1 GDHF01028215 JAI24099.1 GAKP01007130 JAC51822.1 GEDC01011068 JAS26230.1 GFDF01001851 JAV12233.1 KQ978730 KYN28962.1 GL768304 EFZ11532.1 JXJN01004250 JXJN01004251 JXJN01004252 GECU01019743 JAS87963.1 GFDF01001852 JAV12232.1 CCAG010002244 GL445827 EFN89063.1 QOIP01000011 RLU16563.1 KQ976745 KYM75419.1 CP012526 ALC45905.1 KQ460930 KPJ10738.1 KQ981986 KYN31231.1 CP012524 ALC41500.1 KK107021 EZA62321.1 KQ982409 KYQ56670.1 GDKW01001515 JAI55080.1 GL887844 EGI69872.1 GDRN01100499 JAI58557.1 GL442513 EFN63383.1 GEZM01077989 JAV63076.1 GBHO01032077 GBHO01028913 GBRD01001748 GDHC01016007 GDHC01007905 JAG11527.1 JAG14691.1 JAG64073.1 JAQ02622.1 JAQ10724.1 KQ978494 KYM93639.1 KA647027 KA649702 AFP61656.1 APGK01054238 APGK01054239 APGK01054240 KB741248 ENN71949.1 ABLF02039231 KB632305 ERL91852.1 GFTR01004795 JAW11631.1 GGMS01007226 MBY76429.1 NNAY01000606 OXU27415.1 ALC45904.1 GECL01002342 JAP03782.1 SSX13062.1 SSX32502.1 ATLV01020679 KE525304 KFB45540.1 GFXV01001951 MBW13756.1
KPJ03523.1 NWSH01001580 PCG70806.1 KZ149940 PZC76952.1 ODYU01007415 SOQ50158.1 CVRI01000066 CRL06205.1 CH479182 EDW34429.1 CM000070 EAL28619.2 KRT00892.1 GDAI01000130 JAI17473.1 CH902623 EDV30252.1 KPU72753.1 OUUW01000008 SPP84497.1 KQ971343 EFA03243.2 EDW34430.1 GFDL01000508 JAV34537.1 AAAB01008980 EAL39319.2 CH940650 KRF82842.1 CM000160 EDW97021.1 AXCN02001876 KQ414592 KOC70197.1 CH933806 EDW14063.2 KQ761155 OAD58144.1 EDW66649.1 KRF82843.1 KRF82844.1 JRES01001304 KNC23718.1 CH480815 EDW42532.1 CM000364 EDX13295.1 GAPW01002731 JAC10867.1 UFQS01002045 UFQT01002045 SSX13061.1 SSX32501.1 AE014297 AY119214 AAF54725.1 AAM51074.1 AGB95872.1 CH954181 EDV49445.1 DQ440393 ABF18426.1 CH477747 EAT36649.1 KQ435756 KOX75978.1 GEBQ01013191 JAT26786.1 GFDG01001259 JAV17540.1 GGFM01004179 MBW24930.1 GEBQ01025720 JAT14257.1 GGFJ01007436 MBW56577.1 GAMC01013712 GAMC01013711 JAB92843.1 ADMH02000408 ETN66666.1 KA646388 KA647028 AFP61657.1 GGFK01008190 MBW41511.1 KZ288191 PBC34420.1 JR049541 AEY60997.1 GECZ01024041 GECZ01010888 GECZ01010132 JAS45728.1 JAS58881.1 JAS59637.1 ADTU01022691 CH916373 EDV94958.1 GANO01002145 JAB57726.1 GDHF01028215 JAI24099.1 GAKP01007130 JAC51822.1 GEDC01011068 JAS26230.1 GFDF01001851 JAV12233.1 KQ978730 KYN28962.1 GL768304 EFZ11532.1 JXJN01004250 JXJN01004251 JXJN01004252 GECU01019743 JAS87963.1 GFDF01001852 JAV12232.1 CCAG010002244 GL445827 EFN89063.1 QOIP01000011 RLU16563.1 KQ976745 KYM75419.1 CP012526 ALC45905.1 KQ460930 KPJ10738.1 KQ981986 KYN31231.1 CP012524 ALC41500.1 KK107021 EZA62321.1 KQ982409 KYQ56670.1 GDKW01001515 JAI55080.1 GL887844 EGI69872.1 GDRN01100499 JAI58557.1 GL442513 EFN63383.1 GEZM01077989 JAV63076.1 GBHO01032077 GBHO01028913 GBRD01001748 GDHC01016007 GDHC01007905 JAG11527.1 JAG14691.1 JAG64073.1 JAQ02622.1 JAQ10724.1 KQ978494 KYM93639.1 KA647027 KA649702 AFP61656.1 APGK01054238 APGK01054239 APGK01054240 KB741248 ENN71949.1 ABLF02039231 KB632305 ERL91852.1 GFTR01004795 JAW11631.1 GGMS01007226 MBY76429.1 NNAY01000606 OXU27415.1 ALC45904.1 GECL01002342 JAP03782.1 SSX13062.1 SSX32502.1 ATLV01020679 KE525304 KFB45540.1 GFXV01001951 MBW13756.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000183832
UP000008744
+ More
UP000001819 UP000007801 UP000268350 UP000007266 UP000075884 UP000007062 UP000075900 UP000075881 UP000075920 UP000008792 UP000002282 UP000075886 UP000053825 UP000009192 UP000192221 UP000037069 UP000001292 UP000000304 UP000000803 UP000008711 UP000078200 UP000008820 UP000053105 UP000095300 UP000095301 UP000000673 UP000242457 UP000005205 UP000001070 UP000078492 UP000092443 UP000092460 UP000092444 UP000008237 UP000069272 UP000075885 UP000092445 UP000279307 UP000078540 UP000092553 UP000053240 UP000078541 UP000053097 UP000075809 UP000007755 UP000000311 UP000078542 UP000002358 UP000019118 UP000007819 UP000030742 UP000215335 UP000030765
UP000001819 UP000007801 UP000268350 UP000007266 UP000075884 UP000007062 UP000075900 UP000075881 UP000075920 UP000008792 UP000002282 UP000075886 UP000053825 UP000009192 UP000192221 UP000037069 UP000001292 UP000000304 UP000000803 UP000008711 UP000078200 UP000008820 UP000053105 UP000095300 UP000095301 UP000000673 UP000242457 UP000005205 UP000001070 UP000078492 UP000092443 UP000092460 UP000092444 UP000008237 UP000069272 UP000075885 UP000092445 UP000279307 UP000078540 UP000092553 UP000053240 UP000078541 UP000053097 UP000075809 UP000007755 UP000000311 UP000078542 UP000002358 UP000019118 UP000007819 UP000030742 UP000215335 UP000030765
Interpro
Gene 3D
ProteinModelPortal
H9JL47
A0A3S2P8F5
A0A194QDA3
A0A2A4JG98
A0A2W1BPI9
A0A2H1WAS2
+ More
A0A1J1J183 B4GFP4 Q295T9 A0A0K8TTJ3 B3MTX1 A0A3B0JQP7 D6WN36 B4GFP5 A0A1Q3G411 A0A182N8X8 Q5TNS1 A0A182RAG4 A0A182KCQ0 A0A182VSR3 A0A0Q9WEG4 B4PN09 A0A182QU83 A0A0L7RGU2 B4K5M6 A0A310SH91 B4LWS8 A0A1W4UN02 A0A0L0BUP9 B4HHJ3 B4QTJ0 A0A023EN96 A0A336L7W8 Q9VGF7 B3P4D0 A0A1A9VPJ8 Q1HQQ1 Q16QJ1 A0A0N0U648 A0A1B6LSY0 A0A1I8P125 A0A1L8EFP8 A0A2M3Z8W5 A0A1B6KS47 A0A2M4BU31 A0A1I8N795 A0A1I8N794 W8B6Z1 W5JRA3 T1PGN7 A0A2M4AL52 A0A2A3ERV0 V9IK22 A0A1B6G8Z8 A0A158NPX9 B4JT68 U5EUW4 A0A0K8UBS0 A0A034WBY0 A0A1B6DKT8 A0A1L8E0G9 A0A195EM13 E9J6L7 A0A1A9XU33 A0A1B0AVI4 A0A1B6IM18 A0A1L8E0Y7 A0A1B0GCX7 E2B5H7 A0A182F8U5 A0A182PTW5 A0A1A9ZJQ4 A0A3L8D8X7 A0A195ATV9 A0A0M4ET21 A0A194QYX7 A0A195ESU9 A0A0M4EUR8 A0A026X2S8 A0A151X8M8 A0A0N7Z952 F4W7J9 A0A0P4VZG2 E2AT59 A0A1Y1KUJ1 A0A0A9WSU9 A0A151I6B7 T1PE37 K7IMW7 N6SW13 J9JUX6 U4UFS3 A0A224XUK3 A0A2S2QFA0 A0A232F9R8 A0A0M4EDK7 A0A0V0G6W4 A0A336LGC9 A0A084W5P6 A0A2H8TI21
A0A1J1J183 B4GFP4 Q295T9 A0A0K8TTJ3 B3MTX1 A0A3B0JQP7 D6WN36 B4GFP5 A0A1Q3G411 A0A182N8X8 Q5TNS1 A0A182RAG4 A0A182KCQ0 A0A182VSR3 A0A0Q9WEG4 B4PN09 A0A182QU83 A0A0L7RGU2 B4K5M6 A0A310SH91 B4LWS8 A0A1W4UN02 A0A0L0BUP9 B4HHJ3 B4QTJ0 A0A023EN96 A0A336L7W8 Q9VGF7 B3P4D0 A0A1A9VPJ8 Q1HQQ1 Q16QJ1 A0A0N0U648 A0A1B6LSY0 A0A1I8P125 A0A1L8EFP8 A0A2M3Z8W5 A0A1B6KS47 A0A2M4BU31 A0A1I8N795 A0A1I8N794 W8B6Z1 W5JRA3 T1PGN7 A0A2M4AL52 A0A2A3ERV0 V9IK22 A0A1B6G8Z8 A0A158NPX9 B4JT68 U5EUW4 A0A0K8UBS0 A0A034WBY0 A0A1B6DKT8 A0A1L8E0G9 A0A195EM13 E9J6L7 A0A1A9XU33 A0A1B0AVI4 A0A1B6IM18 A0A1L8E0Y7 A0A1B0GCX7 E2B5H7 A0A182F8U5 A0A182PTW5 A0A1A9ZJQ4 A0A3L8D8X7 A0A195ATV9 A0A0M4ET21 A0A194QYX7 A0A195ESU9 A0A0M4EUR8 A0A026X2S8 A0A151X8M8 A0A0N7Z952 F4W7J9 A0A0P4VZG2 E2AT59 A0A1Y1KUJ1 A0A0A9WSU9 A0A151I6B7 T1PE37 K7IMW7 N6SW13 J9JUX6 U4UFS3 A0A224XUK3 A0A2S2QFA0 A0A232F9R8 A0A0M4EDK7 A0A0V0G6W4 A0A336LGC9 A0A084W5P6 A0A2H8TI21
PDB
2C3E
E-value=5.14421e-20,
Score=239
Ontologies
GO
Topology
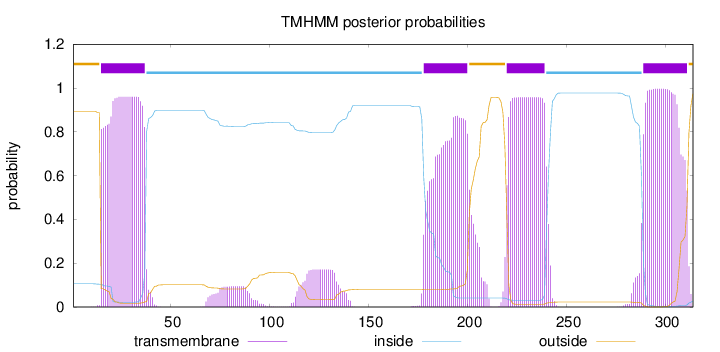
Length:
314
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
88.8694900000001
Exp number, first 60 AAs:
21.47867
Total prob of N-in:
0.10569
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 177
TMhelix
178 - 200
outside
201 - 219
TMhelix
220 - 239
inside
240 - 288
TMhelix
289 - 311
outside
312 - 314
Population Genetic Test Statistics
Pi
270.645407
Theta
214.40423
Tajima's D
0.789463
CLR
0.122557
CSRT
0.593220338983051
Interpretation
Uncertain
