Pre Gene Modal
BGIBMGA002350
Annotation
PREDICTED:_aurora_kinase_B_[Amyelois_transitella]
Full name
Aurora kinase B
+ More
Oxysterol-binding protein
Oxysterol-binding protein
Alternative Name
IPL1/Aurora-like protein kinase
Serine/threonine-protein kinase Ial
Serine/threonine-protein kinase aurora-B
Serine/threonine-protein kinase Ial
Serine/threonine-protein kinase aurora-B
Location in the cell
Cytoplasmic Reliability : 2.477
Sequence
CDS
ATGAAGAGCGAAGTGCTCGAACTTGAAACAAAAATAATAAACCATGATGCCTACGGATCTTCGTACAAGTGGTCTCCAAGAGATTTTGAGCTCGGATCTTCACTAGGCCAAGGCAAATTCGGACATGTTCACGTAGCTAGAGAGAAAAAAACTGGATTCCTTGTTGCCATTAAGACTTTGTTCAAATCTCAAATCGTGAAATCGAAATGTGAACGACAAGTTATGCGAGAAATTGAAATTCAGTCACATTTGAAACACTCAAACATACTTCGACTGCTGACGTGGTTTCATGATGAGAGACGCATATATCTGGTTGTAGAATTTGCAGCTGGTGGAGAACTGTACAAACATCTCACAAATTCACCCCAAGGACGTTTTCCAGAAAGTAAAGCAGCAAGATACATATACCAGGTAGCTGATGCTGTTGAATACTGTCACCAACATCATGTAATACATCGTGACATAAAACCAGAAAATATTCTGGTGGCTTTTAGTGGAGATCTTAAACTTGCTGACTTTGGTTGGTCAGTCCATGCTCCATCTGAAAGACGTAAAACGATGTGTGGAACCCTTGATTATCTGCCACCAGAAATGATAAAACGCGAAGTATATGATGTGTCTGTTGACCACTGGTGCATAGGGGTGTTGCTTTATGAGTTTCTAGTGGGAAAGCCTCCATTTGAAACTGAAGGTGAAGACAAGACATATGCAAGAATATTGAGTCTCGATATTGTATACCCAAGTCATATACCTGATGGAGCCAAGGATTTGATCTCAAAGCTTTTAAGACACTCGAGTAAAGATAGATTGTCCCTTGAAGGAGTAAAAAAACATTATTGGGTCCAACAATTTCAGACAGTATAG
Protein
MKSEVLELETKIINHDAYGSSYKWSPRDFELGSSLGQGKFGHVHVAREKKTGFLVAIKTLFKSQIVKSKCERQVMREIEIQSHLKHSNILRLLTWFHDERRIYLVVEFAAGGELYKHLTNSPQGRFPESKAARYIYQVADAVEYCHQHHVIHRDIKPENILVAFSGDLKLADFGWSVHAPSERRKTMCGTLDYLPPEMIKREVYDVSVDHWCIGVLLYEFLVGKPPFETEGEDKTYARILSLDIVYPSHIPDGAKDLISKLLRHSSKDRLSLEGVKKHYWVQQFQTV
Summary
Description
Serine/threonine-protein kinase which mediates both meiotic and mitotic chromosome segregation. Required for histone H3 'Ser-10' phosphorylation. Phosphorylates mei-S332 within residues 124-126 and stabilizes its association with centromeres during meiosis.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Subunit
Interacts with Incenp and Cdc37.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. Aurora subfamily.
Belongs to the OSBP family.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. Aurora subfamily.
Belongs to the OSBP family.
Keywords
ATP-binding
Cell cycle
Cell division
Chromatin regulator
Chromosome
Chromosome partition
Complete proteome
Cytoplasm
Cytoskeleton
Kinase
Magnesium
Meiosis
Metal-binding
Mitosis
Nucleotide-binding
Reference proteome
Transferase
Feature
chain Aurora kinase B
Uniprot
L8B3S6
A0A2A4J9P7
A0A212EJG1
S4PTA4
A0A194QYZ1
A0A194Q1E1
+ More
H9IYL6 A0A336L5P6 A0A0K8W1X4 A0A034WDS3 W8BXG8 A0A232FLQ2 K7INW3 A0A1W4XRU6 A0A084W3Q3 A0A182W9W9 A0A182GUP7 A0A0A1XP02 Q175B8 A0A182SU91 A0A182L2B7 A0A1S4GWG8 A0A182V3I8 A0A182UKX7 A0A182WTG2 A0A182IEM6 A0A182M4P0 Q7Q3F8 A0A182NLK7 A0A182YPM7 A0A1B0CQ81 E2BXF8 A0A088A1Z6 A0A182R393 A0A1Q3EW20 A0A182IJW2 B4LS72 A0A182R099 W5JR20 A0A0M4EPD2 A0A154PRE7 A0A0L7QUL4 A0A3L8DGR3 B0XL95 A0A182F681 A0A0J7KM67 T1GK03 A0A1B6BZW6 A0A1B0DJA7 B4P132 B4HWX2 B4Q9V4 Q9VKN7 N6UL49 B4KKM4 B4NLN3 Q29PJ0 B4G6W9 A0A0K8V3M6 B3N518 B4JPU6 B3MJX5 A0A3B0JH60 F4X4U4 A0A1B0FH58 A0A1B6FAH1 D6X165 A0A1W4W495 A0A151X625 B4KKM5 A0A2A3ELE1 A0A158NQG8 A0A151IC22 A0A067RI07 A0A1A9UQY8 A0A1B6IYX2 A0A1A9XVS1 A0A1B0ANV9 A0A1Y1L784 A0A1I8PAY1 A0A0C9PH86 A0A2J7QMS1 A0A1I8NFU6 A0A0L0C1S3 A0A1A9WUY1 A0A1I8MLG7 A0A1B6LIQ3 I2B3M3 A0A1J1I9E6 A0A1J1I5M0 R7TMJ4 A7SLH0 A0A0M9AA88 B5THM9 E2AED8 A0A0P5M4A1 A0A0P6FR25 A0A023FWI0 A0A0P5Y7J5
H9IYL6 A0A336L5P6 A0A0K8W1X4 A0A034WDS3 W8BXG8 A0A232FLQ2 K7INW3 A0A1W4XRU6 A0A084W3Q3 A0A182W9W9 A0A182GUP7 A0A0A1XP02 Q175B8 A0A182SU91 A0A182L2B7 A0A1S4GWG8 A0A182V3I8 A0A182UKX7 A0A182WTG2 A0A182IEM6 A0A182M4P0 Q7Q3F8 A0A182NLK7 A0A182YPM7 A0A1B0CQ81 E2BXF8 A0A088A1Z6 A0A182R393 A0A1Q3EW20 A0A182IJW2 B4LS72 A0A182R099 W5JR20 A0A0M4EPD2 A0A154PRE7 A0A0L7QUL4 A0A3L8DGR3 B0XL95 A0A182F681 A0A0J7KM67 T1GK03 A0A1B6BZW6 A0A1B0DJA7 B4P132 B4HWX2 B4Q9V4 Q9VKN7 N6UL49 B4KKM4 B4NLN3 Q29PJ0 B4G6W9 A0A0K8V3M6 B3N518 B4JPU6 B3MJX5 A0A3B0JH60 F4X4U4 A0A1B0FH58 A0A1B6FAH1 D6X165 A0A1W4W495 A0A151X625 B4KKM5 A0A2A3ELE1 A0A158NQG8 A0A151IC22 A0A067RI07 A0A1A9UQY8 A0A1B6IYX2 A0A1A9XVS1 A0A1B0ANV9 A0A1Y1L784 A0A1I8PAY1 A0A0C9PH86 A0A2J7QMS1 A0A1I8NFU6 A0A0L0C1S3 A0A1A9WUY1 A0A1I8MLG7 A0A1B6LIQ3 I2B3M3 A0A1J1I9E6 A0A1J1I5M0 R7TMJ4 A7SLH0 A0A0M9AA88 B5THM9 E2AED8 A0A0P5M4A1 A0A0P6FR25 A0A023FWI0 A0A0P5Y7J5
EC Number
2.7.11.1
Pubmed
27836666
22118469
23622113
26354079
19121390
25348373
+ More
24495485 28648823 20075255 24438588 26483478 25830018 17510324 20966253 12364791 25244985 20798317 17994087 20920257 23761445 30249741 17550304 22936249 10433558 10231582 10731132 12537572 12537569 11266459 11352945 12374737 16824953 23537049 15632085 21719571 18362917 19820115 21347285 24845553 28004739 25315136 26108605 22427657 23254933 17615350
24495485 28648823 20075255 24438588 26483478 25830018 17510324 20966253 12364791 25244985 20798317 17994087 20920257 23761445 30249741 17550304 22936249 10433558 10231582 10731132 12537572 12537569 11266459 11352945 12374737 16824953 23537049 15632085 21719571 18362917 19820115 21347285 24845553 28004739 25315136 26108605 22427657 23254933 17615350
EMBL
KX420635
AB777868
AQQ11601.1
BAM76581.1
NWSH01002306
PCG68569.1
+ More
AGBW02014474 OWR41624.1 GAIX01012383 JAA80177.1 KQ460930 KPJ10753.1 KQ459579 KPI99371.1 BABH01042651 UFQS01001093 UFQT01001093 SSX08970.1 SSX28881.1 GDHF01007156 JAI45158.1 GAKP01007014 GAKP01007011 GAKP01007009 GAKP01007000 JAC51943.1 GAMC01002593 JAC03963.1 NNAY01000039 OXU31664.1 ATLV01020099 KE525294 KFB44847.1 JXUM01089600 JXUM01089601 JXUM01089602 KQ563783 KXJ73311.1 GBXI01001620 JAD12672.1 CH477403 EAT41669.1 AAAB01008964 APCN01005210 AXCM01005979 EAA12168.3 AJWK01023212 GL451254 EFN79606.1 GFDL01015540 JAV19505.1 CH940649 EDW64758.1 AXCN02002185 ADMH02000676 ETN65350.1 CP012523 ALC38687.1 KQ435078 KZC14479.1 KQ414735 KOC62259.1 QOIP01000009 RLU19058.1 DS234366 EDS33747.1 LBMM01005480 KMQ91478.1 CAQQ02019963 GEDC01030492 GEDC01014096 GEDC01013796 JAS06806.1 JAS23202.1 JAS23502.1 AJVK01006302 CM000157 EDW88007.1 CH480818 EDW52517.1 CM000361 CM002910 EDX04621.1 KMY89639.1 AF121358 AF121361 AE014134 AY118562 APGK01019217 KB740096 KB630363 KB631141 KB631630 ENN81411.1 ERL83560.1 ERL84130.1 ERL84863.1 CH933807 EDW12688.1 CH964274 EDW85272.1 CH379058 EAL34303.1 CH479180 EDW28288.1 GDHF01034051 GDHF01032835 GDHF01023976 GDHF01019129 GDHF01006830 GDHF01000903 JAI18263.1 JAI19479.1 JAI28338.1 JAI33185.1 JAI45484.1 JAI51411.1 CH954177 EDV58963.1 CH916372 EDV98926.1 CH902620 EDV32430.1 OUUW01000006 SPP81734.1 GL888679 EGI58517.1 CCAG010021908 GECZ01022567 JAS47202.1 KQ971368 EFA10666.1 KQ982491 KYQ55759.1 EDW12689.2 KZ288215 PBC32623.1 ADTU01023318 KQ978069 KYM97356.1 KK852498 KDR22618.1 GECU01015594 GECU01006637 JAS92112.1 JAT01070.1 JXJN01001068 GEZM01062681 JAV69543.1 GBYB01000213 JAG69980.1 NEVH01013204 PNF29868.1 JRES01001119 KNC25364.1 GEBQ01016412 JAT23565.1 JN021276 AFJ45029.1 CVRI01000042 CRK95590.1 CRK95589.1 AMQN01002464 KB309292 ELT94849.1 DS469698 EDO35437.1 KQ435716 KOX79039.1 EU939755 ACH73237.1 GL438828 EFN68190.1 GDIQ01160836 JAK90889.1 GDIP01063462 GDIQ01046633 GDIQ01046632 JAM40253.1 JAN48104.1 GBBL01001461 JAC25859.1 GDIP01063461 GDIQ01048184 LRGB01001005 JAM40254.1 JAN46553.1 KZS13905.1
AGBW02014474 OWR41624.1 GAIX01012383 JAA80177.1 KQ460930 KPJ10753.1 KQ459579 KPI99371.1 BABH01042651 UFQS01001093 UFQT01001093 SSX08970.1 SSX28881.1 GDHF01007156 JAI45158.1 GAKP01007014 GAKP01007011 GAKP01007009 GAKP01007000 JAC51943.1 GAMC01002593 JAC03963.1 NNAY01000039 OXU31664.1 ATLV01020099 KE525294 KFB44847.1 JXUM01089600 JXUM01089601 JXUM01089602 KQ563783 KXJ73311.1 GBXI01001620 JAD12672.1 CH477403 EAT41669.1 AAAB01008964 APCN01005210 AXCM01005979 EAA12168.3 AJWK01023212 GL451254 EFN79606.1 GFDL01015540 JAV19505.1 CH940649 EDW64758.1 AXCN02002185 ADMH02000676 ETN65350.1 CP012523 ALC38687.1 KQ435078 KZC14479.1 KQ414735 KOC62259.1 QOIP01000009 RLU19058.1 DS234366 EDS33747.1 LBMM01005480 KMQ91478.1 CAQQ02019963 GEDC01030492 GEDC01014096 GEDC01013796 JAS06806.1 JAS23202.1 JAS23502.1 AJVK01006302 CM000157 EDW88007.1 CH480818 EDW52517.1 CM000361 CM002910 EDX04621.1 KMY89639.1 AF121358 AF121361 AE014134 AY118562 APGK01019217 KB740096 KB630363 KB631141 KB631630 ENN81411.1 ERL83560.1 ERL84130.1 ERL84863.1 CH933807 EDW12688.1 CH964274 EDW85272.1 CH379058 EAL34303.1 CH479180 EDW28288.1 GDHF01034051 GDHF01032835 GDHF01023976 GDHF01019129 GDHF01006830 GDHF01000903 JAI18263.1 JAI19479.1 JAI28338.1 JAI33185.1 JAI45484.1 JAI51411.1 CH954177 EDV58963.1 CH916372 EDV98926.1 CH902620 EDV32430.1 OUUW01000006 SPP81734.1 GL888679 EGI58517.1 CCAG010021908 GECZ01022567 JAS47202.1 KQ971368 EFA10666.1 KQ982491 KYQ55759.1 EDW12689.2 KZ288215 PBC32623.1 ADTU01023318 KQ978069 KYM97356.1 KK852498 KDR22618.1 GECU01015594 GECU01006637 JAS92112.1 JAT01070.1 JXJN01001068 GEZM01062681 JAV69543.1 GBYB01000213 JAG69980.1 NEVH01013204 PNF29868.1 JRES01001119 KNC25364.1 GEBQ01016412 JAT23565.1 JN021276 AFJ45029.1 CVRI01000042 CRK95590.1 CRK95589.1 AMQN01002464 KB309292 ELT94849.1 DS469698 EDO35437.1 KQ435716 KOX79039.1 EU939755 ACH73237.1 GL438828 EFN68190.1 GDIQ01160836 JAK90889.1 GDIP01063462 GDIQ01046633 GDIQ01046632 JAM40253.1 JAN48104.1 GBBL01001461 JAC25859.1 GDIP01063461 GDIQ01048184 LRGB01001005 JAM40254.1 JAN46553.1 KZS13905.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000005204
UP000215335
+ More
UP000002358 UP000192223 UP000030765 UP000075920 UP000069940 UP000249989 UP000008820 UP000075901 UP000075882 UP000075903 UP000075902 UP000076407 UP000075840 UP000075883 UP000007062 UP000075884 UP000076408 UP000092461 UP000008237 UP000005203 UP000075900 UP000075880 UP000008792 UP000075886 UP000000673 UP000092553 UP000076502 UP000053825 UP000279307 UP000002320 UP000069272 UP000036403 UP000015102 UP000092462 UP000002282 UP000001292 UP000000304 UP000000803 UP000019118 UP000030742 UP000009192 UP000007798 UP000001819 UP000008744 UP000008711 UP000001070 UP000007801 UP000268350 UP000007755 UP000092444 UP000007266 UP000192221 UP000075809 UP000242457 UP000005205 UP000078542 UP000027135 UP000078200 UP000092443 UP000092460 UP000095300 UP000235965 UP000095301 UP000037069 UP000091820 UP000183832 UP000014760 UP000001593 UP000053105 UP000000311 UP000076858
UP000002358 UP000192223 UP000030765 UP000075920 UP000069940 UP000249989 UP000008820 UP000075901 UP000075882 UP000075903 UP000075902 UP000076407 UP000075840 UP000075883 UP000007062 UP000075884 UP000076408 UP000092461 UP000008237 UP000005203 UP000075900 UP000075880 UP000008792 UP000075886 UP000000673 UP000092553 UP000076502 UP000053825 UP000279307 UP000002320 UP000069272 UP000036403 UP000015102 UP000092462 UP000002282 UP000001292 UP000000304 UP000000803 UP000019118 UP000030742 UP000009192 UP000007798 UP000001819 UP000008744 UP000008711 UP000001070 UP000007801 UP000268350 UP000007755 UP000092444 UP000007266 UP000192221 UP000075809 UP000242457 UP000005205 UP000078542 UP000027135 UP000078200 UP000092443 UP000092460 UP000095300 UP000235965 UP000095301 UP000037069 UP000091820 UP000183832 UP000014760 UP000001593 UP000053105 UP000000311 UP000076858
Interpro
Gene 3D
ProteinModelPortal
L8B3S6
A0A2A4J9P7
A0A212EJG1
S4PTA4
A0A194QYZ1
A0A194Q1E1
+ More
H9IYL6 A0A336L5P6 A0A0K8W1X4 A0A034WDS3 W8BXG8 A0A232FLQ2 K7INW3 A0A1W4XRU6 A0A084W3Q3 A0A182W9W9 A0A182GUP7 A0A0A1XP02 Q175B8 A0A182SU91 A0A182L2B7 A0A1S4GWG8 A0A182V3I8 A0A182UKX7 A0A182WTG2 A0A182IEM6 A0A182M4P0 Q7Q3F8 A0A182NLK7 A0A182YPM7 A0A1B0CQ81 E2BXF8 A0A088A1Z6 A0A182R393 A0A1Q3EW20 A0A182IJW2 B4LS72 A0A182R099 W5JR20 A0A0M4EPD2 A0A154PRE7 A0A0L7QUL4 A0A3L8DGR3 B0XL95 A0A182F681 A0A0J7KM67 T1GK03 A0A1B6BZW6 A0A1B0DJA7 B4P132 B4HWX2 B4Q9V4 Q9VKN7 N6UL49 B4KKM4 B4NLN3 Q29PJ0 B4G6W9 A0A0K8V3M6 B3N518 B4JPU6 B3MJX5 A0A3B0JH60 F4X4U4 A0A1B0FH58 A0A1B6FAH1 D6X165 A0A1W4W495 A0A151X625 B4KKM5 A0A2A3ELE1 A0A158NQG8 A0A151IC22 A0A067RI07 A0A1A9UQY8 A0A1B6IYX2 A0A1A9XVS1 A0A1B0ANV9 A0A1Y1L784 A0A1I8PAY1 A0A0C9PH86 A0A2J7QMS1 A0A1I8NFU6 A0A0L0C1S3 A0A1A9WUY1 A0A1I8MLG7 A0A1B6LIQ3 I2B3M3 A0A1J1I9E6 A0A1J1I5M0 R7TMJ4 A7SLH0 A0A0M9AA88 B5THM9 E2AED8 A0A0P5M4A1 A0A0P6FR25 A0A023FWI0 A0A0P5Y7J5
H9IYL6 A0A336L5P6 A0A0K8W1X4 A0A034WDS3 W8BXG8 A0A232FLQ2 K7INW3 A0A1W4XRU6 A0A084W3Q3 A0A182W9W9 A0A182GUP7 A0A0A1XP02 Q175B8 A0A182SU91 A0A182L2B7 A0A1S4GWG8 A0A182V3I8 A0A182UKX7 A0A182WTG2 A0A182IEM6 A0A182M4P0 Q7Q3F8 A0A182NLK7 A0A182YPM7 A0A1B0CQ81 E2BXF8 A0A088A1Z6 A0A182R393 A0A1Q3EW20 A0A182IJW2 B4LS72 A0A182R099 W5JR20 A0A0M4EPD2 A0A154PRE7 A0A0L7QUL4 A0A3L8DGR3 B0XL95 A0A182F681 A0A0J7KM67 T1GK03 A0A1B6BZW6 A0A1B0DJA7 B4P132 B4HWX2 B4Q9V4 Q9VKN7 N6UL49 B4KKM4 B4NLN3 Q29PJ0 B4G6W9 A0A0K8V3M6 B3N518 B4JPU6 B3MJX5 A0A3B0JH60 F4X4U4 A0A1B0FH58 A0A1B6FAH1 D6X165 A0A1W4W495 A0A151X625 B4KKM5 A0A2A3ELE1 A0A158NQG8 A0A151IC22 A0A067RI07 A0A1A9UQY8 A0A1B6IYX2 A0A1A9XVS1 A0A1B0ANV9 A0A1Y1L784 A0A1I8PAY1 A0A0C9PH86 A0A2J7QMS1 A0A1I8NFU6 A0A0L0C1S3 A0A1A9WUY1 A0A1I8MLG7 A0A1B6LIQ3 I2B3M3 A0A1J1I9E6 A0A1J1I5M0 R7TMJ4 A7SLH0 A0A0M9AA88 B5THM9 E2AED8 A0A0P5M4A1 A0A0P6FR25 A0A023FWI0 A0A0P5Y7J5
PDB
4C2V
E-value=9.17419e-87,
Score=815
Ontologies
GO
GO:0007094
GO:0043988
GO:0004674
GO:0032133
GO:0005524
GO:0032467
GO:0032465
GO:0000780
GO:0051233
GO:0035174
GO:0031616
GO:0005876
GO:0007052
GO:0000776
GO:0048132
GO:0004712
GO:0061952
GO:0030496
GO:0007076
GO:1990385
GO:0006325
GO:0000775
GO:0051321
GO:0046872
GO:0005737
GO:0005819
GO:0006468
GO:0000281
GO:0000070
GO:0030261
GO:0016572
GO:0016021
GO:0004672
GO:0006869
GO:0005634
GO:0005665
GO:0043565
GO:0048384
PANTHER
Topology
Subcellular location
Chromosome
Cytoplasm
Cytoskeleton
Midbody
Cytoplasm
Cytoskeleton
Midbody
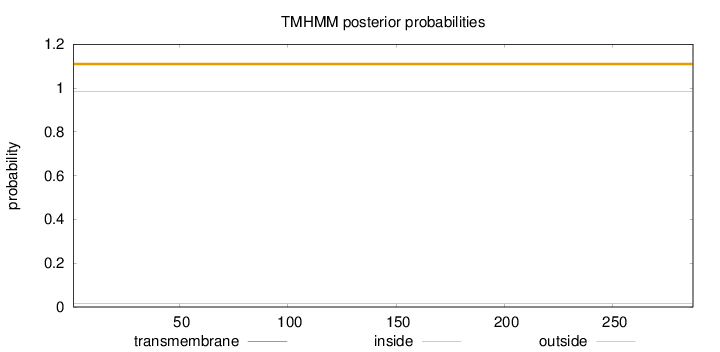
Length:
287
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00208
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.01511
outside
1 - 287
Population Genetic Test Statistics
Pi
235.815548
Theta
197.632801
Tajima's D
0.702549
CLR
0
CSRT
0.577471126443678
Interpretation
Uncertain
