Gene
KWMTBOMO03954
Pre Gene Modal
BGIBMGA010236
Annotation
PREDICTED:_FAM206_family_protein_CG9288_[Amyelois_transitella]
Full name
Protein Abitram
Alternative Name
Actin-binding transcription modulator
Protein Simiate
Protein Simiate
Location in the cell
Nuclear Reliability : 2.199
Sequence
CDS
ATGGATTTCAACATTCTAGATTCAATACCAGACTTAGACAATCTGAAGACCTTCACAGAGAGATACTTTTCAAAACGATACTTAATTAATGTAGATGGCATAAAAAATAATGATATGATGATAATGTTTCATTCAAACCGAATTGCATTGTTATCTCTAGCACCAAGTCATTCCTTCTTCAGTAAAGACGAAAATTACAAAATTAACTTCAACATTGGTAAAATTGATCGTTTGAACAATTCTGTCAAAGGAAAAAGTAAAAAGGGCGGACAACTATTATCTCCAAAATCTGTGATTTGCAAGATAGAATTCAGCGATGAAAGCGTATTTGAAGTGCCCTGTTGCATGAAAGGTACTTTGGTTGAAGTTAATGAAGAACTTGTACAGAATCCATTGTTGCTGAAAAACATGCCAGATTCTGATGGATTTATTGCCATTATGCTAGCAAGTATAGCAATTTCAGAAGCGACAAAAAGTGAGTTGTTAACTCATGAAGAGTACATAAAATGTATTGATTGTTCTGTTAATAAACAATAG
Protein
MDFNILDSIPDLDNLKTFTERYFSKRYLINVDGIKNNDMMIMFHSNRIALLSLAPSHSFFSKDENYKINFNIGKIDRLNNSVKGKSKKGGQLLSPKSVICKIEFSDESVFEVPCCMKGTLVEVNEELVQNPLLLKNMPDSDGFIAIMLASIAISEATKSELLTHEEYIKCIDCSVNKQ
Summary
Description
May regulate actin polymerization.
May regulate actin polymerization, filopodia dynamics and arborization of neurons.
May regulate actin polymerization, filopodia dynamics and arborization of neurons.
Similarity
Belongs to the ABITRAM family.
Keywords
Complete proteome
Reference proteome
Cell projection
Nucleus
Feature
chain Protein Abitram
Uniprot
H9JL35
A0A2A4J9Z8
I4DKC8
S4NT05
A0A2W1BZY7
A0A194R4H5
+ More
A0A212EJI4 A0A0L7LMJ4 A0A1E1WDS2 A0A067QQ89 E2AFK5 A0A0J7KQG0 A0A2J7PY37 E9I8S5 A0A1B6I720 A0A0M9A5Q6 E9H8P1 E2B2R3 A0A2A3EM76 A0A026WSS4 A0A1B6G029 T1JEP8 A0A2T7PBM9 A0A1E1X3V7 A0A2R7WHB2 F4WNK9 A0A0P5YLV3 A0A0P5V7J1 A0A0N7ZX54 R7VL45 K7ITV4 A0A023GAU0 A0A2R5LNF8 A0A293LNN7 A0A232F0W9 A0A0N8EIL6 A0A195ED45 A0A0L7R667 B4NHA8 G3MRZ1 A0A195FW66 A0A0L8FGF0 A0A1S3I8Q1 A0A158NUJ0 A0A0C9RTE5 A0A1E1XVH9 A0A131XGU2 Q1DGE2 A0A1S3HHR9 A0A195B8C7 A7SRF3 A0A131YRZ4 A0A3S3P490 A0A1U7RWL0 Q17GK9 L7M4R5 A0A151IMA2 A0A151NCS3 A0A183J8K0 A0A1S2ZA59 A0A151WGU2 B0X169 B4R0L1 A0A224YZN5 B4HF91 A0A3M6UFE2 A0A1B6C9I2 B3LVS2 A0A023EIT8 A0A0C9RJQ5 B3P400 A0A098M1G1 H3AZ21 Q9VFR1 H2Y7P9 A0A369RSN2 A0A0B8RZG8 B3SC65 A0A3B1J1X9 B4PQJ3 A0A182G5W2 A0A1W4USJ6 A0A3B5KB98 V9LDI1 A0A1X7VV63 A0A2D0PZ28 A0A3Q2NMD2 A0A091DVC0 A0A3Q3K6D9 A0A3Q2NMF3 A0A3Q1G4H4 E0W2B6 A0A3Q2ZIF7 A0A3Q1AN17 A0A3P8TB30 G3H9F6 A0A146ZNE4 Q1LU93 A0A3P8T8J6 V9LCC4
A0A212EJI4 A0A0L7LMJ4 A0A1E1WDS2 A0A067QQ89 E2AFK5 A0A0J7KQG0 A0A2J7PY37 E9I8S5 A0A1B6I720 A0A0M9A5Q6 E9H8P1 E2B2R3 A0A2A3EM76 A0A026WSS4 A0A1B6G029 T1JEP8 A0A2T7PBM9 A0A1E1X3V7 A0A2R7WHB2 F4WNK9 A0A0P5YLV3 A0A0P5V7J1 A0A0N7ZX54 R7VL45 K7ITV4 A0A023GAU0 A0A2R5LNF8 A0A293LNN7 A0A232F0W9 A0A0N8EIL6 A0A195ED45 A0A0L7R667 B4NHA8 G3MRZ1 A0A195FW66 A0A0L8FGF0 A0A1S3I8Q1 A0A158NUJ0 A0A0C9RTE5 A0A1E1XVH9 A0A131XGU2 Q1DGE2 A0A1S3HHR9 A0A195B8C7 A7SRF3 A0A131YRZ4 A0A3S3P490 A0A1U7RWL0 Q17GK9 L7M4R5 A0A151IMA2 A0A151NCS3 A0A183J8K0 A0A1S2ZA59 A0A151WGU2 B0X169 B4R0L1 A0A224YZN5 B4HF91 A0A3M6UFE2 A0A1B6C9I2 B3LVS2 A0A023EIT8 A0A0C9RJQ5 B3P400 A0A098M1G1 H3AZ21 Q9VFR1 H2Y7P9 A0A369RSN2 A0A0B8RZG8 B3SC65 A0A3B1J1X9 B4PQJ3 A0A182G5W2 A0A1W4USJ6 A0A3B5KB98 V9LDI1 A0A1X7VV63 A0A2D0PZ28 A0A3Q2NMD2 A0A091DVC0 A0A3Q3K6D9 A0A3Q2NMF3 A0A3Q1G4H4 E0W2B6 A0A3Q2ZIF7 A0A3Q1AN17 A0A3P8TB30 G3H9F6 A0A146ZNE4 Q1LU93 A0A3P8T8J6 V9LCC4
Pubmed
19121390
22651552
26354079
23622113
28756777
22118469
+ More
26227816 24845553 20798317 21282665 21292972 24508170 30249741 28503490 21719571 23254933 20075255 28648823 17994087 22216098 21347285 26131772 29209593 28049606 17510324 17615350 26830274 25576852 22293439 28797301 30382153 24945155 9215903 10731132 12537572 12537569 30042472 25476704 18719581 25329095 17550304 26483478 21551351 24402279 20566863 21804562 23594743
26227816 24845553 20798317 21282665 21292972 24508170 30249741 28503490 21719571 23254933 20075255 28648823 17994087 22216098 21347285 26131772 29209593 28049606 17510324 17615350 26830274 25576852 22293439 28797301 30382153 24945155 9215903 10731132 12537572 12537569 30042472 25476704 18719581 25329095 17550304 26483478 21551351 24402279 20566863 21804562 23594743
EMBL
BABH01011545
NWSH01002306
PCG68576.1
AK401746
KQ459600
BAM18368.1
+ More
KPI94193.1 GAIX01013837 JAA78723.1 KZ149930 PZC77383.1 KQ460930 KPJ10761.1 AGBW02014474 OWR41618.1 JTDY01000583 KOB76579.1 GDQN01005921 JAT85133.1 KK853141 KDR10756.1 GL439118 EFN67756.1 LBMM01004315 KMQ92573.1 NEVH01020850 PNF21251.1 GL761649 EFZ23020.1 GECU01024987 GECU01009873 JAS82719.1 JAS97833.1 KQ435728 KOX77777.1 GL732605 EFX71850.1 GL445232 EFN90003.1 KZ288219 PBC32269.1 KK107109 QOIP01000008 EZA59082.1 RLU19484.1 GECZ01013993 JAS55776.1 JH432127 PZQS01000005 PVD30819.1 GFAC01005264 JAT93924.1 KK854825 PTY19043.1 GL888237 EGI64202.1 GDIP01055977 LRGB01000084 JAM47738.1 KZS21019.1 GDIP01119213 JAL84501.1 GDIP01203905 JAJ19497.1 AMQN01004070 KB292570 ELU17295.1 GBBM01005613 JAC29805.1 GGLE01006832 MBY10958.1 GFWV01010388 MAA35117.1 NNAY01001370 OXU24209.1 GDIQ01023140 JAN71597.1 KQ979074 KYN22742.1 KQ414648 KOC66329.1 CH964272 EDW84584.1 JO844642 AEO36259.1 KQ981215 KYN44562.1 KQ432350 KOF62783.1 ADTU01002850 GBZX01002119 JAG90621.1 GFAA01000345 JAU03090.1 GEFH01002979 JAP65602.1 CH902418 EAT32221.1 KQ976565 KYM80497.1 DS469761 EDO33701.1 GEDV01006533 JAP82024.1 NCKU01000696 RWS14533.1 CH477258 EAT45790.1 EAT45791.1 GACK01006172 JAA58862.1 KQ977063 KYN05994.1 AKHW03003364 KYO34614.1 UZAM01017183 VDP46300.1 KQ983150 KYQ47037.1 DS232254 EDS38517.1 CM000364 EDX13026.1 GFPF01008074 MAA19220.1 CH480815 EDW42265.1 RCHS01001653 RMX52362.1 GEDC01027187 JAS10111.1 CH902617 EDV43696.1 GAPW01004727 JAC08871.1 GBYB01008430 JAG78197.1 CH954181 EDV49174.1 GBSI01000409 JAC96086.1 AFYH01095442 AFYH01095443 AE014297 AY069582 NOWV01000223 RDD37697.1 GBSH01000506 JAG68518.1 DS985268 EDV19667.1 CM000160 EDW97289.1 JXUM01146730 KQ570231 KXJ68411.1 JW878329 AFP10846.1 KN123095 KFO26721.1 DS235875 EEB19772.1 JH000228 EGV92491.1 GCES01018460 JAR67863.1 BX957315 BX927389 BC125852 JW877803 AFP10320.1
KPI94193.1 GAIX01013837 JAA78723.1 KZ149930 PZC77383.1 KQ460930 KPJ10761.1 AGBW02014474 OWR41618.1 JTDY01000583 KOB76579.1 GDQN01005921 JAT85133.1 KK853141 KDR10756.1 GL439118 EFN67756.1 LBMM01004315 KMQ92573.1 NEVH01020850 PNF21251.1 GL761649 EFZ23020.1 GECU01024987 GECU01009873 JAS82719.1 JAS97833.1 KQ435728 KOX77777.1 GL732605 EFX71850.1 GL445232 EFN90003.1 KZ288219 PBC32269.1 KK107109 QOIP01000008 EZA59082.1 RLU19484.1 GECZ01013993 JAS55776.1 JH432127 PZQS01000005 PVD30819.1 GFAC01005264 JAT93924.1 KK854825 PTY19043.1 GL888237 EGI64202.1 GDIP01055977 LRGB01000084 JAM47738.1 KZS21019.1 GDIP01119213 JAL84501.1 GDIP01203905 JAJ19497.1 AMQN01004070 KB292570 ELU17295.1 GBBM01005613 JAC29805.1 GGLE01006832 MBY10958.1 GFWV01010388 MAA35117.1 NNAY01001370 OXU24209.1 GDIQ01023140 JAN71597.1 KQ979074 KYN22742.1 KQ414648 KOC66329.1 CH964272 EDW84584.1 JO844642 AEO36259.1 KQ981215 KYN44562.1 KQ432350 KOF62783.1 ADTU01002850 GBZX01002119 JAG90621.1 GFAA01000345 JAU03090.1 GEFH01002979 JAP65602.1 CH902418 EAT32221.1 KQ976565 KYM80497.1 DS469761 EDO33701.1 GEDV01006533 JAP82024.1 NCKU01000696 RWS14533.1 CH477258 EAT45790.1 EAT45791.1 GACK01006172 JAA58862.1 KQ977063 KYN05994.1 AKHW03003364 KYO34614.1 UZAM01017183 VDP46300.1 KQ983150 KYQ47037.1 DS232254 EDS38517.1 CM000364 EDX13026.1 GFPF01008074 MAA19220.1 CH480815 EDW42265.1 RCHS01001653 RMX52362.1 GEDC01027187 JAS10111.1 CH902617 EDV43696.1 GAPW01004727 JAC08871.1 GBYB01008430 JAG78197.1 CH954181 EDV49174.1 GBSI01000409 JAC96086.1 AFYH01095442 AFYH01095443 AE014297 AY069582 NOWV01000223 RDD37697.1 GBSH01000506 JAG68518.1 DS985268 EDV19667.1 CM000160 EDW97289.1 JXUM01146730 KQ570231 KXJ68411.1 JW878329 AFP10846.1 KN123095 KFO26721.1 DS235875 EEB19772.1 JH000228 EGV92491.1 GCES01018460 JAR67863.1 BX957315 BX927389 BC125852 JW877803 AFP10320.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000027135 UP000000311 UP000036403 UP000235965 UP000053105 UP000000305 UP000008237 UP000242457 UP000053097 UP000279307 UP000245119 UP000007755 UP000076858 UP000014760 UP000002358 UP000215335 UP000078492 UP000053825 UP000007798 UP000078541 UP000053454 UP000085678 UP000005205 UP000008820 UP000078540 UP000001593 UP000285301 UP000189705 UP000078542 UP000050525 UP000050793 UP000079721 UP000075809 UP000002320 UP000000304 UP000001292 UP000275408 UP000007801 UP000008711 UP000008672 UP000000803 UP000007875 UP000253843 UP000009022 UP000018467 UP000002282 UP000069940 UP000249989 UP000192221 UP000005226 UP000007879 UP000221080 UP000265000 UP000028990 UP000261600 UP000257200 UP000009046 UP000264800 UP000257160 UP000265080 UP000001075 UP000000437
UP000027135 UP000000311 UP000036403 UP000235965 UP000053105 UP000000305 UP000008237 UP000242457 UP000053097 UP000279307 UP000245119 UP000007755 UP000076858 UP000014760 UP000002358 UP000215335 UP000078492 UP000053825 UP000007798 UP000078541 UP000053454 UP000085678 UP000005205 UP000008820 UP000078540 UP000001593 UP000285301 UP000189705 UP000078542 UP000050525 UP000050793 UP000079721 UP000075809 UP000002320 UP000000304 UP000001292 UP000275408 UP000007801 UP000008711 UP000008672 UP000000803 UP000007875 UP000253843 UP000009022 UP000018467 UP000002282 UP000069940 UP000249989 UP000192221 UP000005226 UP000007879 UP000221080 UP000265000 UP000028990 UP000261600 UP000257200 UP000009046 UP000264800 UP000257160 UP000265080 UP000001075 UP000000437
Interpro
Gene 3D
ProteinModelPortal
H9JL35
A0A2A4J9Z8
I4DKC8
S4NT05
A0A2W1BZY7
A0A194R4H5
+ More
A0A212EJI4 A0A0L7LMJ4 A0A1E1WDS2 A0A067QQ89 E2AFK5 A0A0J7KQG0 A0A2J7PY37 E9I8S5 A0A1B6I720 A0A0M9A5Q6 E9H8P1 E2B2R3 A0A2A3EM76 A0A026WSS4 A0A1B6G029 T1JEP8 A0A2T7PBM9 A0A1E1X3V7 A0A2R7WHB2 F4WNK9 A0A0P5YLV3 A0A0P5V7J1 A0A0N7ZX54 R7VL45 K7ITV4 A0A023GAU0 A0A2R5LNF8 A0A293LNN7 A0A232F0W9 A0A0N8EIL6 A0A195ED45 A0A0L7R667 B4NHA8 G3MRZ1 A0A195FW66 A0A0L8FGF0 A0A1S3I8Q1 A0A158NUJ0 A0A0C9RTE5 A0A1E1XVH9 A0A131XGU2 Q1DGE2 A0A1S3HHR9 A0A195B8C7 A7SRF3 A0A131YRZ4 A0A3S3P490 A0A1U7RWL0 Q17GK9 L7M4R5 A0A151IMA2 A0A151NCS3 A0A183J8K0 A0A1S2ZA59 A0A151WGU2 B0X169 B4R0L1 A0A224YZN5 B4HF91 A0A3M6UFE2 A0A1B6C9I2 B3LVS2 A0A023EIT8 A0A0C9RJQ5 B3P400 A0A098M1G1 H3AZ21 Q9VFR1 H2Y7P9 A0A369RSN2 A0A0B8RZG8 B3SC65 A0A3B1J1X9 B4PQJ3 A0A182G5W2 A0A1W4USJ6 A0A3B5KB98 V9LDI1 A0A1X7VV63 A0A2D0PZ28 A0A3Q2NMD2 A0A091DVC0 A0A3Q3K6D9 A0A3Q2NMF3 A0A3Q1G4H4 E0W2B6 A0A3Q2ZIF7 A0A3Q1AN17 A0A3P8TB30 G3H9F6 A0A146ZNE4 Q1LU93 A0A3P8T8J6 V9LCC4
A0A212EJI4 A0A0L7LMJ4 A0A1E1WDS2 A0A067QQ89 E2AFK5 A0A0J7KQG0 A0A2J7PY37 E9I8S5 A0A1B6I720 A0A0M9A5Q6 E9H8P1 E2B2R3 A0A2A3EM76 A0A026WSS4 A0A1B6G029 T1JEP8 A0A2T7PBM9 A0A1E1X3V7 A0A2R7WHB2 F4WNK9 A0A0P5YLV3 A0A0P5V7J1 A0A0N7ZX54 R7VL45 K7ITV4 A0A023GAU0 A0A2R5LNF8 A0A293LNN7 A0A232F0W9 A0A0N8EIL6 A0A195ED45 A0A0L7R667 B4NHA8 G3MRZ1 A0A195FW66 A0A0L8FGF0 A0A1S3I8Q1 A0A158NUJ0 A0A0C9RTE5 A0A1E1XVH9 A0A131XGU2 Q1DGE2 A0A1S3HHR9 A0A195B8C7 A7SRF3 A0A131YRZ4 A0A3S3P490 A0A1U7RWL0 Q17GK9 L7M4R5 A0A151IMA2 A0A151NCS3 A0A183J8K0 A0A1S2ZA59 A0A151WGU2 B0X169 B4R0L1 A0A224YZN5 B4HF91 A0A3M6UFE2 A0A1B6C9I2 B3LVS2 A0A023EIT8 A0A0C9RJQ5 B3P400 A0A098M1G1 H3AZ21 Q9VFR1 H2Y7P9 A0A369RSN2 A0A0B8RZG8 B3SC65 A0A3B1J1X9 B4PQJ3 A0A182G5W2 A0A1W4USJ6 A0A3B5KB98 V9LDI1 A0A1X7VV63 A0A2D0PZ28 A0A3Q2NMD2 A0A091DVC0 A0A3Q3K6D9 A0A3Q2NMF3 A0A3Q1G4H4 E0W2B6 A0A3Q2ZIF7 A0A3Q1AN17 A0A3P8TB30 G3H9F6 A0A146ZNE4 Q1LU93 A0A3P8T8J6 V9LCC4
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus speckle
Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Cell projection Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Lamellipodium Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Nucleus Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Growth cone Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Dendrite Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Cell projection Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Lamellipodium Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Nucleus Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Growth cone Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
Dendrite Localizes to somata and dendrites in cortical neurons. With evidence from 1 publications.
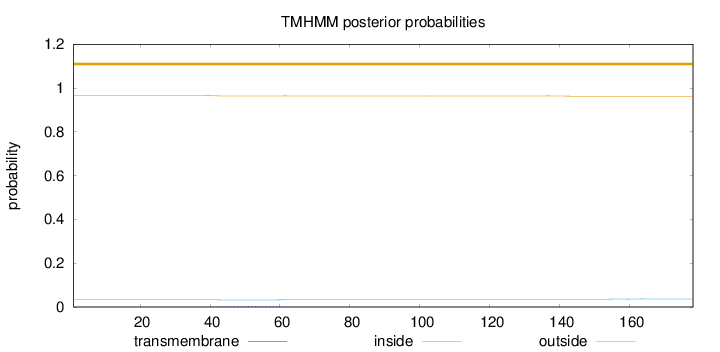
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11415
Exp number, first 60 AAs:
0.08701
Total prob of N-in:
0.03265
outside
1 - 178
Population Genetic Test Statistics
Pi
172.223295
Theta
154.318735
Tajima's D
-0.495853
CLR
92.664024
CSRT
0.24198790060497
Interpretation
Uncertain
